This is a preprint.
Tailoring a CRISPR/Cas-based Epigenome Editor for Programmable Chromatin Acylation and Decreased Cytotoxicity
- PMID: 39345554
- PMCID: PMC11429961
- DOI: 10.1101/2024.09.22.611000
Tailoring a CRISPR/Cas-based Epigenome Editor for Programmable Chromatin Acylation and Decreased Cytotoxicity
Abstract
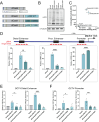
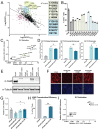
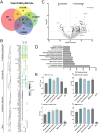
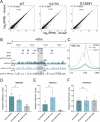
Engineering histone acylation states can inform mechanistic epigenetics and catalyze therapeutic epigenome editing opportunities. Here, we developed engineered lysine acyltransferases that enable the programmable deposition of acetylation and longer-chain acylations. We show that targeting an engineered lysine crotonyltransferase results in weak levels of endogenous enhancer activation yet retains potency when targeted to promoters. We further identify a single mutation within the catalytic core of human p300 that preserves enzymatic activity while substantially reducing cytotoxicity, enabling improved viral delivery. We leveraged these capabilities to perform single-cell CRISPR activation screening and map enhancers to the genes they regulate in situ. We also discover acylation-specific interactions and find that recruitment of p300, regardless of catalytic activity, to prime editing sites can improve editing efficiency. These new programmable epigenome editing tools and insights expand our ability to understand the mechanistic role of lysine acylation in epigenetic and cellular processes and perform functional genomic screens.
Keywords: CRISPR; CRISPRa; EP300; Epigenome editing; Perturb-seq; enhancer; histone acetylation; histone crotonylation; off-target editing; protein engineering.
Conflict of interest statement
DECLARATION OF INTERESTS J.G. and I.B.H are inventors on patents related to this work. J.G., J.L., B.M., M.E., and I.B.H. are inventors on patents related to genome and epigenome editing technologies. J.G. and I.B.H. are founders of Mercator Biosciences.
Figures


References
-
- Goell J. H. & Hilton I. B. CRISPR/Cas-Based Epigenome Editing: Advances, Applications, and Clinical Utility. Trends in Biotechnology 39, 678–691 (2021). - PubMed
-
- Nakamura M., Gao Y., Dominguez A. A. & Qi L. S. CRISPR technologies for precise epigenome editing. Nat Cell Biol 23, 11–22 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
