This is a preprint.
Regulatory logic and transposable element dynamics in nematode worm genomes
- PMID: 39345564
- PMCID: PMC11429677
- DOI: 10.1101/2024.09.15.613132
Regulatory logic and transposable element dynamics in nematode worm genomes
Abstract
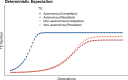
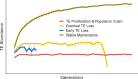
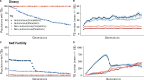
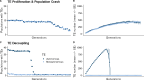
Genome sequencing has revealed a tremendous diversity of transposable elements (TEs) in eukaryotes but there is little understanding of the evolutionary processes responsible for TE diversity. Non-autonomous TEs have lost the machinery necessary for transposition and rely on closely related autonomous TEs for critical proteins. We studied two mathematical models of TE regulation, one assuming that both autonomous tranposons and their non-autonomous relatives operate under the same regulatory logic, competing for transposition resources, and one assuming that autonomous TEs self-attenuate transposition while non-autonomous transposons continually increase, parasitizing their autonomous relatives. We implemented these models in stochastic simulations and studied how TE regulatory relationships influence transposons and populations. We found that only outcrossing populations evolving with Parasitic TE regulation resulted in stable maintenance of TEs. We tested our model predictions in Caenorhabditis genomes by annotating TEs in two focal families, autonomous LINEs and their non-autonomous SINE relatives and the DNA transposon Mutator. We found broad variation in autonomous - non-autonomous relationships and rapid mutational decay in the sequences that allow non-autonomous TEs to transpose. Together, our results suggest that individual TE families evolve according to disparate regulatory rules that are relevant in the early, acute stages of TE invasion.
Conflict of interest statement
Competing interests No competing interest is declared.
Figures



Similar articles
-
Species-specific chromatin landscape determines how transposable elements shape genome evolution.Elife. 2022 Aug 23;11:e81567. doi: 10.7554/eLife.81567. Elife. 2022. PMID: 35997258 Free PMC article.
-
A transposable element annotation pipeline and expression analysis reveal potentially active elements in the microalga Tisochrysis lutea.BMC Genomics. 2018 May 22;19(1):378. doi: 10.1186/s12864-018-4763-1. BMC Genomics. 2018. PMID: 29783941 Free PMC article.
-
Diversity and evolution of transposable elements in the plant-parasitic nematodes.BMC Genomics. 2024 May 23;25(1):511. doi: 10.1186/s12864-024-10435-7. BMC Genomics. 2024. PMID: 38783171 Free PMC article.
-
A Field Guide to Eukaryotic Transposable Elements.Annu Rev Genet. 2020 Nov 23;54:539-561. doi: 10.1146/annurev-genet-040620-022145. Epub 2020 Sep 21. Annu Rev Genet. 2020. PMID: 32955944 Free PMC article. Review.
-
Transposable element evolution in plant genome ecosystems.Curr Opin Plant Biol. 2023 Oct;75:102418. doi: 10.1016/j.pbi.2023.102418. Epub 2023 Jul 15. Curr Opin Plant Biol. 2023. PMID: 37459733 Review.
References
-
- Arkhipova I. and Meselson M.. Deleterious transposable elements and the extinction of asexuals. BioEssays, 27:76–85, 2005. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
