This is a preprint.
The transcriptomic landscape of spinal V1 interneurons reveals a role for En1 in specific elements of motor output
- PMID: 39345580
- PMCID: PMC11429899
- DOI: 10.1101/2024.09.18.613279
The transcriptomic landscape of spinal V1 interneurons reveals a role for En1 in specific elements of motor output
Abstract
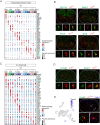
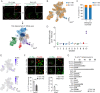
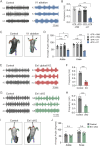
Neural circuits in the spinal cord are composed of diverse sets of interneurons that play crucial roles in shaping motor output. Despite progress in revealing the cellular architecture of the spinal cord, the extent of cell type heterogeneity within interneuron populations remains unclear. Here, we present a single-nucleus transcriptomic atlas of spinal V1 interneurons across postnatal development. We find that the core molecular taxonomy distinguishing neonatal V1 interneurons perdures into adulthood, suggesting conservation of function across development. Moreover, we identify a key role for En1, a transcription factor that marks the V1 population, in specifying one unique subset of V1Pou6f2 interneurons. Loss of En1 selectively disrupts the frequency of rhythmic locomotor output but does not disrupt flexion/extension limb movement. Beyond serving as a molecular resource for this neuronal population, our study highlights how deep neuronal profiling provides an entry point for functional studies of specialized cell types in motor output.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
