This is a preprint.
Population genomics of Plasmodium ovale species in sub-Saharan Africa
- PMID: 39345628
- PMCID: PMC11429939
- DOI: 10.1101/2024.04.10.588912
Population genomics of Plasmodium ovale species in sub-Saharan Africa
Update in
-
Population genomics of Plasmodium ovale species in sub-Saharan Africa.Nat Commun. 2024 Nov 27;15(1):10297. doi: 10.1038/s41467-024-54667-3. Nat Commun. 2024. PMID: 39604397 Free PMC article.
Abstract
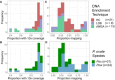
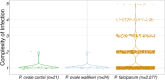
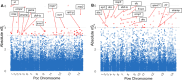
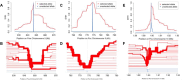
Plasmodium ovale curtisi (Poc) and Plasmodium ovale wallikeri (Pow) are relapsing malaria parasites endemic to Africa and Asia that were previously thought to represent a single species. Amid increasing detection of ovale malaria in sub-Saharan Africa, we performed a population genomic study of both species across the continent. We conducted whole-genome sequencing of 25 isolates from Central and East Africa and analyzed them alongside 20 previously published African genomes. Isolates were predominantly monoclonal (43/45), with their genetic similarity aligning with geography. Pow showed lower average nucleotide diversity (1.8×10-4) across the genome compared to Poc (3.0×10-4) (p < 0.0001). Signatures of selective sweeps involving the dihydrofolate reductase gene were found in both species, as were signs of balancing selection at the merozoite surface protein 1 gene. Differences in the nucleotide diversity of Poc and Pow may reflect unique demographic history, even as similar selective forces facilitate their resilience to malaria control interventions.
Conflict of interest statement
Competing Interests: J.B.P. reports research support from Gilead Sciences, non-financial support from Abbott Laboratories, and consulting for Zymeron Corporation, all outside the scope of this study. The remaining authors declare no competing interests.
Figures


Similar articles
-
Population genomics of Plasmodium ovale species in sub-Saharan Africa.Nat Commun. 2024 Nov 27;15(1):10297. doi: 10.1038/s41467-024-54667-3. Nat Commun. 2024. PMID: 39604397 Free PMC article.
-
Genetic Diversity and Phylogenetic Relatedness of Plasmodium ovale curtisi and Plasmodium ovale wallikeri in sub-Saharan Africa.Microorganisms. 2022 Jun 2;10(6):1147. doi: 10.3390/microorganisms10061147. Microorganisms. 2022. PMID: 35744665 Free PMC article.
-
New reference genomes to distinguish the sympatric malaria parasites, Plasmodium ovale curtisi and Plasmodium ovale wallikeri.Sci Rep. 2024 Feb 15;14(1):3843. doi: 10.1038/s41598-024-54382-5. Sci Rep. 2024. PMID: 38360879 Free PMC article.
-
Comparison of Plasmodium ovale curtisi and Plasmodium ovale wallikeri infections by a meta-analysis approach.Sci Rep. 2021 Mar 19;11(1):6409. doi: 10.1038/s41598-021-85398-w. Sci Rep. 2021. PMID: 33742015 Free PMC article.
-
Two sympatric types of Plasmodium ovale and discrimination by molecular methods.J Microbiol Immunol Infect. 2017 Oct;50(5):559-564. doi: 10.1016/j.jmii.2016.08.004. Epub 2016 Dec 18. J Microbiol Immunol Infect. 2017. PMID: 28065415 Review.
References
-
- Geneva: World Health Organization. World Malaria Report 2023. vol. Licence: CC BY-NC-SA 3.0 IGO. (2023).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
