This is a preprint.
Complete Genomic Characterization of Global Pathogens, Respiratory Syncytial Virus (RSV), and Human Norovirus (HuNoV) Using Probe-based Capture Enrichment
- PMID: 39345650
- PMCID: PMC11429736
- DOI: 10.1101/2024.09.16.613242
Complete Genomic Characterization of Global Pathogens, Respiratory Syncytial Virus (RSV), and Human Norovirus (HuNoV) Using Probe-based Capture Enrichment
Update in
-
Complete genomic characterization of global pathogens respiratory syntical virus and human norovirus using probe based capture enrichment.Sci Rep. 2025 Jul 1;15(1):20526. doi: 10.1038/s41598-025-03398-6. Sci Rep. 2025. PMID: 40594045 Free PMC article.
Abstract
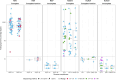
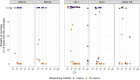
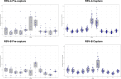
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in children worldwide, while human noroviruses (HuNoV) are a leading cause of epidemic and sporadic acute gastroenteritis. Generating full-length genome sequences for these viruses is crucial for understanding viral diversity and tracking emerging variants. However, obtaining high-quality sequencing data is often challenging due to viral strain variability, quality, and low titers. Here, we present a set of comprehensive oligonucleotide probe sets designed from 1,570 RSV and 1,376 HuNoV isolate sequences in GenBank. Using these probe sets and a capture enrichment sequencing workflow, 85 RSV positive nasal swab samples and 55 (49 stool and six human intestinal enteroids) HuNoV positive samples encompassing major subtypes and genotypes were characterized. The Ct values of these samples ranged from 17.0-29.9 for RSV, and from 20.2-34.8 for HuNoV, with some HuNoV having below the detection limit. The mean percentage of post-processing reads mapped to viral genomes was 85.1% for RSV and 40.8% for HuNoV post-capture, compared to 0.08% and 1.15% in pre-capture libraries, respectively. Full-length genomes were>99% complete in all RSV positive samples and >96% complete in 47/55 HuNoV positive samples-a significant improvement over genome recovery from pre-capture libraries. RSV transcriptome (subgenomic mRNAs) sequences were also characterized from this data. Probe-based capture enrichment offers a comprehensive approach for RSV and HuNoV genome sequencing and monitoring emerging variants.
Keywords: Respiratory syncytial virus (RSV); capture enrichment; genome sequencing; human norovirus (HuNoV).
Figures


References
-
- Li Y, Wang X, Blau DM, Caballero MT, Feikin DR, Gill CJ, Madhi SA, Omer SB, Simoes EAF, Campbell H, Pariente AB, Bardach D, Bassat Q, Casalegno JS, Chakhunashvili G, Crawford N, Danilenko D, Do LAH, Echavarria M, Gentile A, Gordon A, Heikkinen T, Huang QS, Jullien S, Krishnan A, Lopez EL, Markic J, Mira-Iglesias A, Moore HC, Moyes J, Mwananyanda L, Nokes DJ, Noordeen F, Obodai E, Palani N, Romero C, Salimi V, Satav A, Seo E, Shchomak Z, Singleton R, Stolyarov K, Stoszek SK, von Gottberg A, Wurzel D, Yoshida LM, Yung CF, Zar HJ, Respiratory Virus Global Epidemiology N, Nair H, et al. 2022. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet 399:2047–2064. - PMC - PubMed
-
- Yen C, Wikswo ME, Lopman BA, Vinje J, Parashar UD, Hall AJ. 2011. Impact of an emergent norovirus variant in 2009 on norovirus outbreak activity in the United States. Clin Infect Dis 53:568–71. - PubMed
-
- Pangesti KNA, Abd El Ghany M, Walsh MG, Kesson AM, Hill-Cawthorne GA. 2018. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 28. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
