Exploring Structure-Activity Relationships and Modes of Action of Laterocidine
- PMID: 39345814
- PMCID: PMC11428279
- DOI: 10.1021/acscentsci.4c00776
Exploring Structure-Activity Relationships and Modes of Action of Laterocidine
Abstract
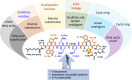
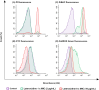
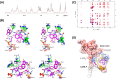
A significant increase in life-threatening infections caused by Gram-negative "superbugs" is a serious threat to global health. With a dearth of new antibiotics in the developmental pipeline, antibiotics with novel mechanisms of action are urgently required to prevent a return to the preantibiotic era. A key strategy to develop novel anti-infective treatments is to discover new natural scaffolds with distinct mechanisms of action. Laterocidine is a unique cyclic lipodepsipeptide with activity against multiple problematic multidrug-resistant Gram-negative pathogens, including Pseudomonas aeruginosa, Acinetobacter baumannii, and Enterobacterales. Here, we developed a total chemical synthesis methodology for laterocidine and undertook systematic structure-activity relationship studies with chemical biology and NMR. We discovered important structural features that drive the antimicrobial activity of laterocidine, leading to the discovery of an engineered peptide surpassing the efficacy of the original peptide. This engineered peptide demonstrated complete inhibition of the growth of a polymyxin-resistant strain of Pseudomonas aeruginosa in static time-kill experiments.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.mBio. 2022 Jun 28;13(3):e0036722. doi: 10.1128/mbio.00367-22. Epub 2022 Apr 14. mBio. 2022. PMID: 35420470 Free PMC article.
-
Large-scale combination screens reveal small-molecule sensitization of antibiotic-resistant gram-negative ESKAPE pathogens.Proc Natl Acad Sci U S A. 2025 Apr;122(13):e2402017122. doi: 10.1073/pnas.2402017122. Epub 2025 Mar 24. Proc Natl Acad Sci U S A. 2025. PMID: 40127266
-
Enhancing colistin efficacy with combination therapies for multidrug-resistant P. aeruginosa and A. baumannii isolates.Future Microbiol. 2025 May-Jun;20(7-9):523-531. doi: 10.1080/17460913.2025.2490377. Epub 2025 Apr 10. Future Microbiol. 2025. PMID: 40208781
-
Macrolide antibiotics (including azithromycin) for cystic fibrosis.Cochrane Database Syst Rev. 2024 Feb 27;2(2):CD002203. doi: 10.1002/14651858.CD002203.pub5. Cochrane Database Syst Rev. 2024. PMID: 38411248 Free PMC article.
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
Cited by
-
Organic Synthesis and Catalysis Enable Facile Access to Bioactive Compounds and Drugs.ACS Cent Sci. 2024 Dec 16;11(1):1-5. doi: 10.1021/acscentsci.4c02041. eCollection 2025 Jan 22. ACS Cent Sci. 2024. PMID: 39866703 Free PMC article. No abstract available.
-
A novel approach for the synthesis of the cyclic lipopeptide globomycin.RSC Med Chem. 2024 Oct 21;16(1):373-8. doi: 10.1039/d4md00685b. Online ahead of print. RSC Med Chem. 2024. PMID: 39493230 Free PMC article.
References
-
- O’Neill J.Antimicrobial resistance: tackling a crisis for the health and wealth of nations; Wellcome Collection, 2015; p 1.
-
- WHO priority pathogens list for R&D of new antibiotics. World Health Organization 2017, https://www.who.int/news/item/17-05-2024-who-updates-list-of-drug-resist....
Grants and funding
LinkOut - more resources
Full Text Sources
