Pan-Cancer Screening and Validation of CALU's Role in EMT Regulation and Tumor Microenvironment in Triple-Negative Breast Cancer
- PMID: 39345892
- PMCID: PMC11439346
- DOI: 10.2147/JIR.S477846
Pan-Cancer Screening and Validation of CALU's Role in EMT Regulation and Tumor Microenvironment in Triple-Negative Breast Cancer
Abstract
Purpose: Cancer-associated fibroblasts (CAFs) significantly contribute to tumor progression and the development of resistance to therapies across a range of malignancies, notably breast cancer. This study aims to elucidate the specific role and prognostic relevance of CALU across multiple cancer types.
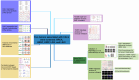
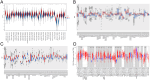
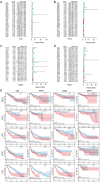
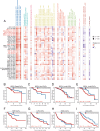
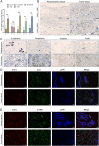
Patients and methods: The association between CALU expression and prognosis, along with clinical characteristics in BRCA, HNSC, KIRP, LGG, and LIHC, was analyzed using data from the TCGA, GTEx, and GEO databases. Transcriptomic analysis of TCGA BRCA project data provided insights into the interaction between CALU and epithelial-mesenchymal transition (EMT) marker genes. Using TIMER and TISCH databases, the correlation between CALU expression and tumor microenvironment infiltration was assessed, alongside an evaluation of CALU expression across various cell types. Furthermore, CALU's influence on TNBC BRCA cell lines was explored, and its expression in tumor tissues was confirmed through immunohistochemical analysis of clinical samples.
Results: This study revealed a consistent upregulation of CALU across several tumor types, including BRCA, KIRP, LIHC, HNSC, and LGG, with elevated CALU expression being associated with unfavorable prognoses. CALU expression was particularly enhanced in clinical contexts linked to poor outcomes. Genomic analysis identified copy number alterations as the principal factor driving CALU overexpression. Additionally, a positive correlation between CALU expression and CAF infiltration was observed, along with its involvement in the EMT process in both CAFs and malignant cells. In vitro experiments demonstrated that CALU is highly expressed in TNBC-BRCA cell lines, and knockdown of CALU effectively reversed EMT progression and inhibited cellular migration. Immunohistochemical analysis of clinical samples corroborated the elevated expression of CALU in tumors, along with alterations in EMT markers.
Conclusion: This comprehensive pan-cancer analysis underscores CALU's critical role in modulating the tumor microenvironment and facilitating cell migration via the EMT pathway, identifying it as a potential therapeutic target.
Keywords: CALU; breast cancer; cancer-associated fibroblasts; epithelial-mesenchymal transition; pan-cancer analysis; prognosis.
© 2024 Chen et al.
Conflict of interest statement
The authors declare no financial non-financial or competing interests.
Figures






References
LinkOut - more resources
Full Text Sources

