Biological roles of THRAP3, STMN1 and GNA13 in human blood cancer cells
- PMID: 39345963
- PMCID: PMC11424602
- DOI: 10.1007/s13205-024-04093-5
Biological roles of THRAP3, STMN1 and GNA13 in human blood cancer cells
Abstract
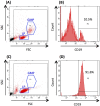
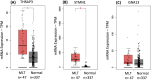
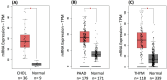
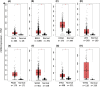
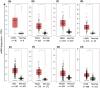
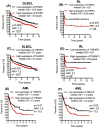
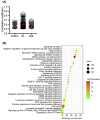
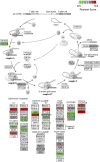
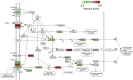
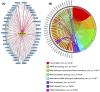
Blood cancers, such as diffuse large B-cell lymphoma (DLBCL), Burkitt's lymphoma (BL) and acute myeloid leukemia (AML), are aggressive neoplasms that are characterized by undesired clinical courses with dismal survival rates. The objective of the current work is to study the expression THRAP3, STMN1 and GNA13 in DLBCL, BL and AML, and to investigate if these proteins are implicated in the prognosis and progression of the blood cancers. Isolation of normal blood cells was performed using lymphoprep coupled with gradient centrifugation and magnetic beads. Flow-cytometric analysis showed high quality of the isolated cells. Western blotting identified THRAP3, STMN1 and GNA13 to be overexpressed in the blood cancer cells but hardly detected in normal blood cells from healthy donors. Consistently, investigations performed using genotype-tissue expression (GTEx) and gene expression profiling interactive analysis (GEPIA) showed that the three proteins had higher mRNA expression in various cancers compared with matched normal tissues (p ≤ 0.01). Furthermore, the up-regulated transcript expression of these proteins was a feature of short overall survival (OS; p ≤ 0.02) in patients with the blood cancers. Interestingly, functional profiling using gProfiler and protein-protein interaction network analysis using STRING with cytoscape reported THRAP3 to be associated with cancer-dependent proliferation and survival pathways (corrected p ≤ 0.05) and to interact with proteins (p = 1 × 10-16) implicated in tumourigenesis and chemotherapy resistance. Taken together, these findings indicated a possible implication of THRAP3, STMN1 and GNA13 in the progression and prognosis of the blood cancers. Additional work using clinical samples of the blood cancers is required to further investigate and validate the results reported here.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-024-04093-5.
Keywords: Blood cancers; GNA13; STMN1; THRAP3.
© King Abdulaziz City for Science and Technology 2024. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestsThe author has no competing interests to declare that are relevant to the content of this article.
Figures

References
-
- Alsagaby SA, Khanna S, Hart KW, Pratt G, Fegan C, Pepper C, Brewis IA, Brennan P (2014) Proteomics-based strategies to identify proteins relevant to chronic lymphocytic leukemia. J Proteome Res 13:5051–5062 - PubMed
-
- Alsagaby SA, Brewis IA, Vijayakumar R, Alhumaydhi FA, Alwashmi AS, Alharbi NK, Al AW, Premanathan M, Pratt G, Fegan C (2021) Proteomics-based identification of cancer-associated proteins in chronic lymphocytic leukaemia. Electron J Biotechnol 52:1–12
LinkOut - more resources
Full Text Sources
Miscellaneous

