The Characteristic of Biofilm Formation in ESBL-Producing K. pneumoniae Isolates
- PMID: 39346024
- PMCID: PMC11427726
- DOI: 10.1155/2024/1802115
The Characteristic of Biofilm Formation in ESBL-Producing K. pneumoniae Isolates
Abstract
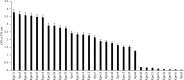
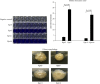
Klebsiella pneumoniae is a pathogen that commonly causes hospital-acquired infections. Bacterial biofilms are structured bacterial communities that adhere to the surface of objects or biological tissues. In this study, we investigated the genome homology and biofilm formation capacity of ESBL-producing K. pneumoniae. Thirty ESBL-producing K. pneumoniae isolates from 25 inpatients at Ruijin Hospital, Shanghai, were subjected to pulsed-field gel electrophoresis (PFGE) to estimate genomic relatedness. Based on the chromosomal DNA patterns we obtained, we identified 21 PFGE profiles from the 30 isolates, eight of which had high homology indicating that they may have genetic relationships and/or potential clonal advantages within the hospital. Approximately 84% (21/25) of the clinical patients had a history of surgery, urinary tract catheterization, and/or arteriovenous intubation, all of which may have increased the risk for nosocomial infections. Biofilms were observed in 73% (22/30) of the isolates and that strains did not express type 3 fimbriae did not have biofilm formation capacity. Above findings indicated that a high percentage of ESBL-producing K. pneumoniae isolates formed biofilms in vitro and even though two strains with cut-off of PFGE reached 100% similarity, they generated biofilms differently. Besides, the variability in biofilm formation ability may be correlated with the expression of type 3 fimbriae. Thus, we next screened four ESBL-producing K. pneumoniae isolates (Kpn5, Kpn7, Kpn11, and Kpn16) with high homology and significant differences in biofilm formation using PFGE molecular typing, colony morphology, and crystal violet tests. Kpn7 and Kpn16 had stronger biofilm formation abilities compared with Kpn5 and Kpn11. The ability of above four ESBL-producing K. pneumoniae isolates to agglutinate in a mannose-resistant manner or in a mannose-sensitive manner, as well as RNA sequencing-based transcriptome results, showed that type 3 fimbriae play a significant role in biofilm formation. In contrast, type 1 fimbriae were downregulated during biofilm formation. Further research is needed to fully understand the regulatory mechanisms which underlie these processes.
Copyright © 2024 Xiaofang Gao et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




Similar articles
-
Characterization of a multidrug-resistant Klebsiella pneumoniae ST3330 clone responsible for a nosocomial outbreak in a neonatal intensive care unit.Ann Palliat Med. 2020 May;9(3):1092-1102. doi: 10.21037/apm-20-958. Epub 2020 May 14. Ann Palliat Med. 2020. PMID: 32434364
-
Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation.BMC Microbiol. 2010 Jun 23;10:179. doi: 10.1186/1471-2180-10-179. BMC Microbiol. 2010. PMID: 20573190 Free PMC article.
-
The Determination and Correlation of Various Virulence Genes, ESBL, Serum Bactericidal Effect and Biofilm Formation of Clinical Isolated Classical Klebsiella pneumoniae and Hypervirulent Klebsiella pneumoniae from Respiratory Tract Infected Patients.Pol J Microbiol. 2017 Dec 4;66(4):501-508. doi: 10.5604/01.3001.0010.7042. Pol J Microbiol. 2017. PMID: 29319515
-
Biotyping, virulotyping and biofilm formation ability of ESBL-Klebsiella pneumoniae isolates from nosocomial infections.J Appl Microbiol. 2022 Jun;132(6):4555-4568. doi: 10.1111/jam.15563. Epub 2022 Apr 13. J Appl Microbiol. 2022. PMID: 35384170
-
Strain-specific transmission in an outbreak of ESBL-producing Enterobacteriaceae in the hemato-oncology care unit: a cohort study.BMC Infect Dis. 2017 Jan 5;17(1):26. doi: 10.1186/s12879-016-2144-4. BMC Infect Dis. 2017. PMID: 28056827 Free PMC article.
Cited by
-
Antibiotic Susceptibility Patterns and Virulence Profiles of Classical and Hypervirulent Klebsiella pneumoniae Strains Isolated from Clinical Samples in Khyber Pakhtunkhwa, Pakistan.Pathogens. 2025 Jan 15;14(1):79. doi: 10.3390/pathogens14010079. Pathogens. 2025. PMID: 39861040 Free PMC article.
References
-
- Hsu C., Chang I., Hsieh P., et al. A novel role for the Klebsiella pneumoniae sap (sensitivity to antimicrobial peptides) transporter in intestinal cell interactions, innate immune responses, liver abscess, and virulence. The Journal of Infectious Diseases . 2019;219(8):1294–1306. doi: 10.1093/infdis/jiy615. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

