A new species of alien land flatworm in the Southern United States
- PMID: 39346042
- PMCID: PMC11430170
- DOI: 10.7717/peerj.17904
A new species of alien land flatworm in the Southern United States
Abstract
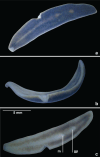
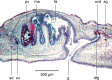
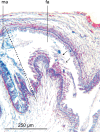
Specimens of a flat and dark brown land planarian were found in a plant nursery in North Carolina, USA in 2020. On the basis of examination of photographs of the live specimens only, the specimens were considered as belonging to Obama nungara, a species originally from South America, which has now invaded a large part of Europe. Unexpectedly, a molecular analysis revealed that the specimens did not belong to this species, neither to the genus Obama. We then undertook its histological study, which finally confirmed that the species is a member of the genus Amaga: the species is herein described as a new species, Amaga pseudobama n. sp. The species has been found in three locations in North Carolina and some infested plants were from Georgia. We reinvestigated specimens collected in Florida in 2015 and found that they also belong to this species. Citizen science observations suggest its presence in other states. Therefore, it is likely that A. pseudobama has already invaded a part of south-east USA and that the invasion took place more than ten years ago. The complete 14,909 bp long mitochondrial genome was obtained. The mitogenome is colinear with those of other Geoplanidae and it was possible to find and annotate a tRNA-Thr, which has been reported missing in several geoplanids. Amaga pseudobama shares with other Geoplaninae the presence of alternative start codons in three protein-coding genes of its mitogenome. The availability of this new genome helped us to improve our annotations of the ND3 gene, for which an ATT start codon is now suggested. Also, the sequence of the ATP6 gene raised questions concerning the use of genetic code 9 to translate the protein-coding genes of Geoplanidae, as the whole translated protein would not contain a single methionine residue when using this code. Two maximum likelihood phylogenies were obtained from genomic data. The first one was based on concatenated alignments of the partial 28S, Elongation Factor 1-alpha (EF1) and cox1 genes. The second was obtained from a concatenated alignment of the mitochondrial proteins. Both strictly discriminate A. pseudobama from O. nungara and instead associate it with Amaga expatria. We note that the nine species currently accepted within Amaga can be separated into two groups, one with extrabulbar prostatic apparatus, including the type species A. amagensis, and one with intrabulbar prostatic apparatus, including the new species A. pseudobama. This suggests that species of the latter group should be separated from Amaga and constitute a new genus. This finding again illustrates the possible emergence of new invasive species in regions naturally devoid of large land planarians, such as North America. Amaga pseudobama thus deserves to be monitored in the USA, although its superficial resemblance to O. nungara and Geoplana arkalabamensis will complicate the use of photographs obtained from citizen science. Our molecular information provides tools for this monitoring.
Keywords: Invasive alien species; Land flatworms; Mitogenome; New species; Platyhelminthes; Taxonomy; USA.
©2024 Justine et al.
Conflict of interest statement
Jean-Lou Justine is an Academic Editor for PeerJ.
Figures












References
-
- Almeida AL, Álvarez-Presas M, Carbayo F. The discovery of new Chilean taxa revolutionizes the systematics of Geoplaninae Neotropical land planarians (Platyhelminthes: Tricladida) Zoological Journal of the Linnean Society. 2023;197:837–898. doi: 10.1093/zoolinnean/zlac072. - DOI
-
- Almeida AL, Francoy TM, Álvarez-Presas M, Carbayo F. Convergent evolution: a new subfamily for bipaliin-like Chilean land planarians (platyhelminthes) Zoologica Scripta. 2021;50:500–508. doi: 10.1111/zsc.12479. - DOI
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

