RNA sequencing identifies lung cancer lineage and facilitates drug repositioning
- PMID: 39346064
- PMCID: PMC11430167
- DOI: 10.7717/peerj.18159
RNA sequencing identifies lung cancer lineage and facilitates drug repositioning
Abstract
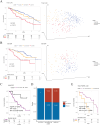
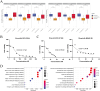
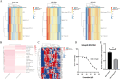
Recent breakthrough therapies have improved survival rates in non-small cell lung cancer (NSCLC), but a paradigm for prospective confirmation is still lacking. Patientdatasets were mainly downloaded from TCGA, CPTAC and GEO. We conducted downstream analysis by collecting metagenes and generated 42-gene subtype classifiers to elucidate biological pathways. Subsequently, scRNA, eRNA, methylation, mutation, and copy number variation were depicted from a phenotype perspective. Enhancing the clinical translatability of molecular subtypes, preclinical models including CMAP, CCLE, and GDSC were utilized for drug repositioning. Importantly, we verified the presence of previously described three phenotypes including bronchioid, neuroendocrine, and squamoid. Poor prognosis was seen in squamoid and neuroendocrine clusters for treatment-naive and immunotherapy populations. The neuroendocrine cluster was dominated by STK11 mutations and 14q13.3 amplifications, whose related methylated loci are predictive of immunotherapy. And the greatest therapeutic potential lies in the bronchioid cluster. We further estimated the relative cell abundance of the tumor microenvironment (TME), specific cell types could be reflected among three clusters. Meanwhile, the higher portion of immune cell infiltration belonged to bronchioid and squamoid, not the neuroendocrine cluster. In drug repositioning, MEK inhibitors resisted bronchioid but were squamoid-sensitive. To conceptually validate compounds/targets, we employed RNA-seq and CCK-8/western blot assays. Our results indicated that dinaciclib and alvocidib exhibited similar activity and sensitivity in the neuroendocrine cluster. Also, a lineage factor named KLF5 recognized by inferred transcriptional factors activity could be suppressed by verteporfin.
Keywords: Drug sensitivity; Lung adenocarcinoma; Metagene; Molecular classification.
© 2024 Zeng et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nature Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. - DOI - PMC - PubMed
-
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. - DOI - PMC - PubMed
-
- Bischoff P, Trinks A, Obermayer B, Pett JP, Wiederspahn J, Uhlitz F, Liang X, Lehmann A, Jurmeister P, Elsner A, Dziodzio T, Rückert JC, Neudecker J, Falk C, Beule D, Sers C, Morkel M, Horst D, Blüthgen N, Klauschen F. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene. 2021;40(50):6748–6758. doi: 10.1038/s41388-021-02054-3. - DOI - PMC - PubMed
-
- Cai L, Lin S, Girard L, Zhou Y, Yang L, Ci B, Zhou Q, Luo D, Yao B, Tang H, Allen J, Huffman K, Gazdar A, Heymach J, Wistuba I, Xiao G, Minna J, Xie Y. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38(14):2551–2564. doi: 10.1038/s41388-018-0588-2. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

