Identification of a novel immune infiltration-related gene signature, MCEMP1, for coronary artery disease
- PMID: 39346078
- PMCID: PMC11438437
- DOI: 10.7717/peerj.18135
Identification of a novel immune infiltration-related gene signature, MCEMP1, for coronary artery disease
Abstract
Background: This study aims to identify a novel gene signature for coronary artery disease (CAD), explore the role of immune cell infiltration in CAD pathogenesis, and assess the cell function of mast cell-expressed membrane protein 1 (MCEMP1) in human umbilical vein endothelial cells (HUVECs) treated with oxidized low-density lipoprotein (ox-LDL).
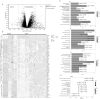
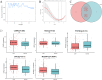
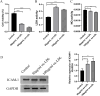
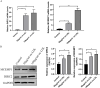
Methods: To identify differentially expressed genes (DEGs) of CAD, datasets GSE24519 and GSE61145 were downloaded from the Gene Expression Omnibus (GEO) database using the R "limma" package with p < 0.05 and |log2 FC| > 1. Gene ontology (GO) and pathway analyses were conducted to determine the biological functions of DEGs. Hub genes were identified using support vector machine-recursive feature elimination (SVM-RFE) and least absolute shrinkage and selection operator (LASSO). The expression levels of these hub genes in CAD were validated using the GSE113079 dataset. CIBERSORT program was used to quantify the proportion of immune cell infiltration. Western blot assay and qRT-PCR were used to detect the expression of hub genes in ox-LDL-treated HUVECs to validate the bioinformatics results. Knockdown interference sequences for MCEMP1 were synthesized, and cell proliferation and apoptosis were examined using a CCK8 kit and Muse® Cell Analyzer, respectively. The concentrations of IL-1β, IL-6, and TNF-α were measured with respective enzyme-linked immunosorbent assay (ELISA) kits.
Results: A total of 73 DEGs (four down-regulated genes and 69 up-regulated genes) were identified in the metadata (GSE24519 and GSE61145) cohort. GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis results indicated that these DEGs might be associated with the regulation of platelet aggregation, defense response or response to bacterium, NF-kappa B signaling pathway, and lipid and atherosclerosis. Using SVM-RFE and LASSO, seven hub genes were obtained from the metadata. The upregulated expression of DIRC2 and MCEMP1 in CAD was confirmed in the GSE113079 dataset and in ox-LDL-treated HUVECs. The associations between the two hub genes (DIRC2 and MCEMP1) and the 22 types of immune cell infiltrates in CAD were found. MCEMP1 knockdown accelerated cell proliferation and suppressed cell apoptosis for ox-LDL-treated HUVECs. Additionally, MCEMP1 knockdown appeared to decrease the expression of inflammatory factors IL-1β, IL-6, and TNF-α.
Conclusions: The results of this study indicate that MCEMP1 may play an important role in CAD pathophysiology.
Keywords: Coronary artery disease; Immune cell infiltration; MCEMP1; Machine learning algorithm; ox-LDL treatment HUVECs cells.
© 2024 Ye et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Six potential biomarkers in septic shock: a deep bioinformatics and prospective observational study.Front Immunol. 2023 Jun 8;14:1184700. doi: 10.3389/fimmu.2023.1184700. eCollection 2023. Front Immunol. 2023. PMID: 37359526 Free PMC article.
-
Identification of hub genes and their correlation with immune infiltration in coronary artery disease through bioinformatics and machine learning methods.J Thorac Dis. 2022 Jul;14(7):2621-2634. doi: 10.21037/jtd-22-632. J Thorac Dis. 2022. PMID: 35928610 Free PMC article.
-
In Silico Identification of Key Genes and Immune Infiltration Characteristics in Epicardial Adipose Tissue from Patients with Coronary Artery Disease.Biomed Res Int. 2022 Oct 29;2022:5610317. doi: 10.1155/2022/5610317. eCollection 2022. Biomed Res Int. 2022. PMID: 36345357 Free PMC article.
-
ZNF429 Participates in the Progression of Coronary Heart Disease through Regulating Inflammatory and Adhesive Factors.Front Biosci (Landmark Ed). 2024 Sep 24;29(9):335. doi: 10.31083/j.fbl2909335. Front Biosci (Landmark Ed). 2024. PMID: 39344313
-
JUN and ATF3 in Gout: Ferroptosis-related potential diagnostic biomarkers.Heliyon. 2024 Oct 30;10(22):e39957. doi: 10.1016/j.heliyon.2024.e39957. eCollection 2024 Nov 30. Heliyon. 2024. PMID: 39619595 Free PMC article. Review.
References
-
- Bampatsias D, Mavroeidis I, Tual-Chalot S, Vlachogiannis NI, Bonini F, Sachse M, Mavraganis G, Mareti A, Kritsioti C, Laina A, Delialis D, Ciliberti G, Sopova K, Gatsiou A, Martelli F, Georgiopoulos G, Stellos K, Stamatelopoulos K. Beta-secretase-1 antisense RNA is associated with vascular ageing and atherosclerotic cardiovascular disease. Thrombosis and Haemostasis. 2022;122(11):1932–1942. doi: 10.1055/a-1914-2094. - DOI - PMC - PubMed
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

