Developing an enhanced chimeric permuted intron-exon system for circular RNA therapeutics
- PMID: 39346546
- PMCID: PMC11426236
- DOI: 10.7150/thno.98214
Developing an enhanced chimeric permuted intron-exon system for circular RNA therapeutics
Abstract
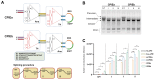
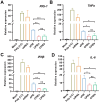
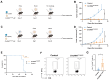
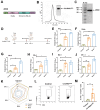
Rationale: Circular RNA (circRNA) therapeutics hold great promise as an iteration strategy in messenger RNA (mRNA) therapeutics due to their inherent stability and durable protein translation capability. Nevertheless, the efficiency of RNA circularization remains a significant constraint, particularly in establishing large-scale manufacturing processes for producing highly purified circRNAs. Hence, it is imperative to develop a universal and more efficient RNA circularization system when considering synthetic circRNAs as therapeutic agents with prospective clinical applications. Methods: We initially developed a chimeric RNA circularization system based on the original permuted intron-exon (PIE) and subsequently established a high-performance liquid chromatography (HPLC) method to obtain highly purified circRNAs. We then evaluated their translational ability and immunogenicity. The circRNAs expressing human papillomavirus (HPV) E7 peptide (43-62aa) and dimerized receptor binding domain (dRBD) from SARS-CoV-2 were encapsulated within lipid nanoparticles (LNPs) as vaccines, followed by an assessment of the in vivo efficacy through determination of antigen-specific T and B cell responses, respectively. Results: We have successfully developed a universal chimeric permuted intron-exon system (CPIE) through engineering of group I self-splicing introns derived from Anabaena pre-tRNALeu or T4 phage thymidylate (Td) synthase gene. Within CPIE, we have effectively enhanced RNA circularization efficiency. By utilizing size exclusion chromatography, circRNAs were effectively separated, which exhibit low immunogenicity and sustained potent protein expression property. In vivo data demonstrate that the constructed circRNA vaccines can elicit robust immune activation (B cell and/or T cell responses) against tumor or SARS-CoV-2 and its variants in mouse models. Conclusions: Overall, we provide an efficient and universal system to synthesize circRNA in vitro, which has extensive application prospect for circRNA therapeutics.
Keywords: chimeric permuted intron-exon system (CPIE); circular RNA (circRNA); mRNA therapeutics; ribozyme; size exclusion chromatography.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Rohner E, Yang R, Foo KS, Goedel A, Chien KR. Unlocking the promise of mRNA therapeutics. Nat Biotechnol. 2022;40:1586–600. - PubMed
-
- Tang P, Hasan MR, Chemaitelly H, Yassine HM, Benslimane FM, Al Khatib HA. et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat Med. 2021;27:2136–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

