Bulk and single-cell transcriptomics identify gene signatures of stem cell-derived NK cell donors with superior cytolytic activity
- PMID: 39346765
- PMCID: PMC11426129
- DOI: 10.1016/j.omton.2024.200870
Bulk and single-cell transcriptomics identify gene signatures of stem cell-derived NK cell donors with superior cytolytic activity
Abstract
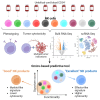
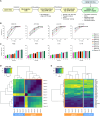
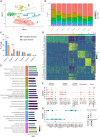
Allogeneic natural killer (NK) cell therapies are a valuable treatment option for cancer, given their remarkable safety and favorable efficacy profile. Although the use of allogeneic donors allows for off-the-shelf and timely patient treatment, intrinsic interindividual differences put clinical efficacy at risk. The identification of donors with superior anti-tumor activity is essential to ensure the success of adoptive NK cell therapies. Here, we investigated the heterogeneity of 10 umbilical cord blood stem cell-derived NK cell batches. First, we evaluated the donors' cytotoxic potential against tumor cell lines from solid and hematological cancer indications, to distinguish a group of superior, "excellent" killers (4/10), compared with "good" killers (6/10). Next, bulk and single-cell RNA sequencing, performed at different stages of NK differentiation, revealed distinct transcriptomic features of the two groups. Excellent donors showed an enrichment in cytotoxicity pathways and a depletion of myeloid traits, linked to the presence of a larger population of effector-like NK cells early on during differentiation. Consequently, we defined a multi-factorial gene expression signature able to predict the donors' cytotoxic potential. Our study contributes to the identification of key traits of superior NK cell batches, supporting the development of efficacious NK therapeutics and the achievement of durable anti-tumor responses.
Keywords: MT: Regular Issue; NK cells; NK cytotoxicity; NK functionality; allogeneic NK cells; cell therapy; donor selection; hematopoietic stem cells; omics; scRNA-seq; umbilical cord blood.
© 2024 The Author(s).
Conflict of interest statement
A.A.v.V., D.S., A.D.D., A.-M.G., J.S., and M.R. are employees of Glycostem Therapeutics. All authors declare no competing interests. Data and results generated, shared, and reported in conjunction with this publication have been filed in a patent application by Glycostem Therapeutics.
Figures



References
-
- Pende D., Parolini S., Pessino A., Sivori S., Augugliaro R., Morelli L., Marcenaro E., Accame L., Malaspina A., Biassoni R., et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
