Unraveling the Molecular Landscape of SCN1A Gene Knockout in Cerebral Organoids: A Multiomics Approach Utilizing Proteomics, Lipidomics, and Transcriptomics
- PMID: 39346820
- PMCID: PMC11425820
- DOI: 10.1021/acsomega.4c05039
Unraveling the Molecular Landscape of SCN1A Gene Knockout in Cerebral Organoids: A Multiomics Approach Utilizing Proteomics, Lipidomics, and Transcriptomics
Abstract
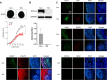
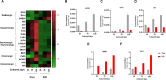
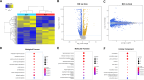
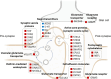
This study investigates the impact of sodium channel protein type 1 subunit alpha (SCN1A) gene knockout (SCN1A KO) on brain development and function using cerebral organoids coupled with a multiomics approach. From comprehensive omics analyses, we found that SCN1A KO organoids exhibit decreased growth, dysregulated neurotransmitter levels, and altered lipidomic, proteomic, and transcriptomic profiles compared to controls under matrix-free differentiation conditions. Neurochemical analysis reveals reduced levels of key neurotransmitters, and lipidomic analysis highlights changes in ether phospholipids and sphingomyelin. Furthermore, quantitative profiling of the SCN1A KO organoid proteome shows perturbations in cholesterol metabolism and sodium ion transportation, potentially affecting synaptic transmission. These findings suggest dysregulation of cholesterol metabolism and sodium ion transport, with implications for synaptic transmission. Overall, these insights shed light on the molecular mechanisms underlying SCN1A-associated disorders, such as Dravet syndrome, and offer potential avenues for therapeutic intervention.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Miller I. O.; Sotero de Menezes M. A.. SCN1A Seizure Disorders. In GeneReviews; Adam M. P.; Feldman J.; Mirzaa G. M.; Pagon R. A.; Wallace S. E.; Bean L. J.; Gripp K. W.; Amemiya A., Eds.; University of Washington: Seattle: Seattle (WA), 1993. - PubMed
-
- Valassina N.; Brusco S.; Salamone A.; Serra L.; Luoni M.; Giannelli S.; Bido S.; Massimino L.; Ungaro F.; Mazzara P. G.; D’Adamo P.; Lignani G.; Broccoli V.; Colasante G. Scn1a Gene Reactivation after Symptom Onset Rescues Pathological Phenotypes in a Mouse Model of Dravet Syndrome. Nat. Commun. 2022, 13 (1), 161 10.1038/s41467-021-27837-w. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
