Causal relationship, shared genes between rheumatoid arthritis and pulp and periapical disease: evidence from GWAS and transcriptome data
- PMID: 39346909
- PMCID: PMC11427265
- DOI: 10.3389/fimmu.2024.1440753
Causal relationship, shared genes between rheumatoid arthritis and pulp and periapical disease: evidence from GWAS and transcriptome data
Abstract
Objective: Patients with rheumatoid arthritis (RA) have an increased risk of developing pulp and periapical disease (PAP), but the causal relationship and shared genetic factors between these conditions have not been explored. This study aimed to investigate the bidirectional causal relationship between RA and PAP and to analyze shared genes and pathogenic pathways.
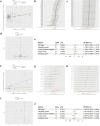
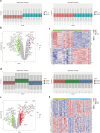
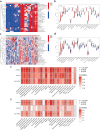
Methods: We utilized GWAS data from the IEU Open GWAS Project and employed five Mendelian randomization methods (MR Egger, weighted median, inverse variance weighted, simple mode, and weighted mode) to investigate the bidirectional causal relationship between RA and PAP. Transcriptome data for RA and irreversible pulpitis (IRP) were obtained from the GEO database. Hub genes were identified through differential analysis, CytoHubba, machine learning (ML), and other methods. The immune infiltration of both diseases was analyzed using the ssGSEA method. Finally, we constructed a regulatory network for miRNAs, transcription factors, chemicals, diseases, and RNA-binding proteins based on the identified hub genes.
Results: RA was significantly associated with an increased risk of PAP (OR = 1.1284, 95% CI 1.0674-1.1929, p < 0.001). However, there was insufficient evidence to support the hypothesis that PAP increased the risk of RA. Integrating datasets and differential analysis identified 84 shared genes primarily involved in immune and inflammatory pathways, including the IL-17 signaling pathway, Th17 cell differentiation, and TNF signaling pathway. Using CytoHubba and three ML methods, we identified three hub genes (HLA-DRA, ITGAX, and PTPRC) that are significantly correlated and valuable for diagnosing RA and IRP. We then constructed a comprehensive regulatory network using the miRDB, miRWalk, ChipBase, hTFtarget, CTD, MalaCards, DisGeNET, and ENCORI databases.
Conclusion: RA may increase the risk of PAP. The three key genes, HLA-DRA, ITGAX, and PTPRC, have significant diagnostic value for both RA and IRP.
Keywords: Mendelian randomization; hub genes; irreversible pulpitis; pulp and periapical disease; rheumatoid arthritis.
Copyright © 2024 Wu, Wang and Qiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

