Rapid PRRSV-2 ORF5-based lineage classification using Nextclade
- PMID: 39346961
- PMCID: PMC11427352
- DOI: 10.3389/fvets.2024.1419340
Rapid PRRSV-2 ORF5-based lineage classification using Nextclade
Abstract
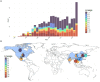
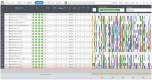
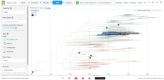
Porcine reproductive and respiratory syndrome virus (PRRSV) continues to be a global challenge for swine health. Yim-Im et al. 2023 provides a standard genetic nomenclature, extending previously published works to better characterize PRRSV-2 ORF5-based genetic lineages on a global scale. To facilitate the use of this nomenclature, scaffold sequences, including historical and contemporary vaccines, were synthesized into a dataset designed for Nextclade v3.0. Metadata from the scaffold sequences representing year, country, and RFLP typing of the sequence were incorporated into the dataset. These scaffold sequences were processed through the Augur pipeline using DQ478308.1 as a reference strain for rooting and comparison. The resultant classifier can be accessed through the Nextclade website (https://clades.nextstrain.org/) or a link on the PRRSView homepage (https://prrsv.vdl.iastate.edu/). The resultant classifier functions the same as other classifiers hosted by the Nextclade core group and can provide phylogenetic-based PRRSV-2 ORF5 classifications on demand. Nextclade provides additional sequence metrics such as classification quality and notable mutations relative to the reference. The submitted sequences are grafted to the reference tree using phylogenetic placement, allowing for comparison to nearby sequences of reference viruses and vaccine strains. Additional comparisons between sequences can be made with metadata incorporated in the dataset. Although Nextclade is hosted as a webtool, the sequences are not uploaded to a server, and all analysis stay strictly confidential to the user. This work provides a standardized, trivial workflow facilitated by Nextclade to rapidly assign lineage classifications to PRRSV-2, identify mutations of interest, and compare contemporary strains to relevant vaccines.
Keywords: ORF5; PRRSV; classification; lineage; sublineage.
Copyright © 2024 Zeller, Chang, Trevisan, Main, Gauger and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Holtkamp D., Rotto H., Garcia R.. (2007). The economic cost of major health challenges in large U.S. swine production systems. American Association of Swine Veterinarians. Orlando, FL. 85–89.
-
- Zimmerman JJ, Dee SA, Holtkamp DJ, Murtaugh MP, Stadejek T, Stevenson GW, et al. Porcine reproductive and respiratory syndrome viruses (porcine arteriviruses) Diseases of swine. (2019). 685–708.
LinkOut - more resources
Full Text Sources

