Capturing Autoinhibited PDK1 Reveals the Linker's Regulatory Role, Informing Innovative Inhibitor Design
- PMID: 39348509
- PMCID: PMC12101721
- DOI: 10.1021/acs.jcim.4c01392
Capturing Autoinhibited PDK1 Reveals the Linker's Regulatory Role, Informing Innovative Inhibitor Design
Abstract
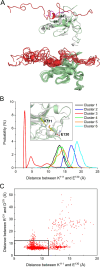
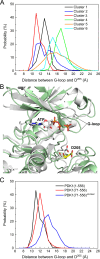
PDK1 is crucial for PI3K/AKT/mTOR and Ras/MAPK cancer signaling. It phosphorylates AKT in a PIP3-dependent but S6K, SGK, and RSK kinases in a PIP3-independent manner. Unlike its substrates, its autoinhibited monomeric state has been unclear, likely due to its low population time, and phosphorylation in the absence of PIP3 has been puzzling too. Here, guided by experimental data, we constructed models and performed all-atom molecular dynamics simulations. In the autoinhibited PDK1 conformation that resembles autoinhibited AKT, binding of the linker between the kinase and PH domains to the PIF-binding pocket promotes the formation of the Glu130-Lys111 salt bridge and weakens the association of the kinase domain with the PH domain, shifting the population from the autoinhibited state to states accessible to the membrane and its kinase substrates. The interaction of the substrates' hydrophobic motif and the PDK1 PIF-binding pocket facilitates the release of the autoinhibition even in the absence of PIP3. Phosphorylation of the serine-rich motif within the linker further attenuates the association of the PH domain with the kinase domain. These suggest that while the monomeric autoinhibited state is relatively stable, it can readily shift to its active, catalysis-prone state to phosphorylate its diverse substrates. Our findings reveal the PDK1 activation mechanism and discover the regulatory role of PDK1's linker, which lead to two innovative linker-based inhibitor strategies: (i) locking the autoinhibited PDK1 through optimization of the interactions of AKT inhibitors with the PH domain of PDK1 and (ii) analogs (small molecules or peptidomimetics) that mimic the linker interactions with the PIF-binding pocket.
Figures





Similar articles
-
The choreography of protein kinase PDK1 and its diverse substrate dance partners.Biochem J. 2023 Oct 11;480(19):1503-1532. doi: 10.1042/BCJ20220396. Biochem J. 2023. PMID: 37792325 Review.
-
Activation of the essential kinase PDK1 by phosphoinositide-driven trans-autophosphorylation.Nat Commun. 2022 Apr 6;13(1):1874. doi: 10.1038/s41467-022-29368-4. Nat Commun. 2022. PMID: 35387990 Free PMC article.
-
The PH domain of phosphoinositide-dependent kinase-1 exhibits a novel, phospho-regulated monomer-dimer equilibrium with important implications for kinase domain activation: single-molecule and ensemble studies.Biochemistry. 2013 Jul 16;52(28):4820-9. doi: 10.1021/bi400488f. Epub 2013 Jul 9. Biochemistry. 2013. PMID: 23745598 Free PMC article.
-
Akt is efficiently activated by PIF-pocket- and PtdIns(3,4,5)P3-dependent mechanisms leading to resistance to PDK1 inhibitors.Biochem J. 2012 Dec 1;448(2):285-95. doi: 10.1042/BJ20121287. Biochem J. 2012. PMID: 23030823
-
Dissecting the role of the 3-phosphoinositide-dependent protein kinase-1 (PDK1) signalling pathways.Cell Cycle. 2008 Oct;7(19):2978-82. doi: 10.4161/cc.7.19.6810. Epub 2008 Oct 18. Cell Cycle. 2008. PMID: 18802401 Review.
Cited by
-
ERK Allosteric Activation: The Importance of Two Ordered Phosphorylation Events.J Mol Biol. 2025 Apr 9:169130. doi: 10.1016/j.jmb.2025.169130. Online ahead of print. J Mol Biol. 2025. PMID: 40216017
-
Uncovering the Role of Distal Regions in PDK1 Allosteric Activation.ACS Bio Med Chem Au. 2025 Mar 24;5(2):299-309. doi: 10.1021/acsbiomedchemau.5c00025. eCollection 2025 Apr 16. ACS Bio Med Chem Au. 2025. PMID: 40255282 Free PMC article.
-
The mechanism of oncogenic PI3K lipid kinase variants at the membrane and their cryptic pockets.bioRxiv [Preprint]. 2025 Jun 29:2025.06.26.661751. doi: 10.1101/2025.06.26.661751. bioRxiv. 2025. PMID: 40666967 Free PMC article. Preprint.
References
-
- Cordon-Barris L., Pascual-Guiral S., Yang S., Gimenez-Llort L., Lope-Piedrafita S., Niemeyer C., Claro E., Lizcano J. M., Bayascas J. R.. Mutation of the 3-Phosphoinositide-Dependent Protein Kinase 1 (PDK1) Substrate-Docking Site in the Developing Brain Causes Microcephaly with Abnormal Brain Morphogenesis Independently of Akt, Leading to Impaired Cognition and Disruptive Behaviors. Mol. Cell. Biol. 2016;36:2967–2982. doi: 10.1128/MCB.00230-16. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

