TCA metabolism regulates DNA hypermethylation in LPS and Mycobacterium tuberculosis-induced immune tolerance
- PMID: 39348545
- PMCID: PMC11474056
- DOI: 10.1073/pnas.2404841121
TCA metabolism regulates DNA hypermethylation in LPS and Mycobacterium tuberculosis-induced immune tolerance
Abstract
Severe and chronic infections, including pneumonia, sepsis, and tuberculosis (TB), induce long-lasting epigenetic changes that are associated with an increase in all-cause postinfectious morbidity and mortality. Oncology studies identified metabolic drivers of the epigenetic landscape, with the tricarboxylic acid (TCA) cycle acting as a central hub. It is unknown if the TCA cycle also regulates epigenetics, specifically DNA methylation, after infection-induced immune tolerance. The following studies demonstrate that lipopolysaccharide and Mycobacterium tuberculosis induce changes in DNA methylation that are mediated by the TCA cycle. Infection-induced DNA hypermethylation is mitigated by inhibitors of cellular metabolism (rapamycin, everolimus, metformin) and the TCA cycle (isocitrate dehydrogenase inhibitors). Conversely, exogenous supplementation with TCA metabolites (succinate and itaconate) induces DNA hypermethylation and immune tolerance. Finally, TB patients who received everolimus have less DNA hypermethylation demonstrating proof of concept that metabolic manipulation can mitigate epigenetic scars.
Keywords: DNA methylation; Rheostat; immune tolerance; sepsis; tuberculosis.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

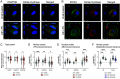
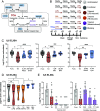
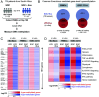
Comment in
-
Linking metabolic and epigenetic changes in immune tolerance.Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2419751121. doi: 10.1073/pnas.2419751121. Epub 2024 Nov 6. Proc Natl Acad Sci U S A. 2024. PMID: 39503896 Free PMC article. No abstract available.
References
-
- Hmama Z., Pena-Diaz S., Joseph S., Av-Gay Y., Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol. Rev. 264, 220–232 (2015). - PubMed
MeSH terms
Substances
Grants and funding
- 1R01AI70774-01A1/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01AG078268/HHS | NIH | NIAID | Division of Intramural Research (DIR, NIAID)
- AI141681-04/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI137527/AI/NIAID NIH HHS/United States
- D43TW012274/HHS | NIH | Fogarty International Center (FIC)
- R01AI137527/HHS | NIH | NIAID | Division of Intramural Research (DIR, NIAID)
- U01GH00227847/HHS | NIH | NHLBI | Division of Intramural Research (DIR)
- P30ES030285 1P42ES0327725/HHS | NIH | National Institute of Environmental Health Sciences (NIEHS)
- 1R01AG078268-01A1/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- OPP1127276-2015/Bill and Melinda Gates Foundation (GF)
- R01 AG078268/AG/NIA NIH HHS/United States
- W81XWH1910026/DOD | OSD | Defense Technical Information Center (DTIC)
- K23 AI141681/AI/NIAID NIH HHS/United States
- R01 AI170774/AI/NIAID NIH HHS/United States
- P50 MD015496/MD/NIMHD NIH HHS/United States
- D43 TW012274/TW/FIC NIH HHS/United States
- RP210227/Cancer Prevention and Research Institute of Texas (CPRIT)
- R01AI165721/HHS | NIH | NIAID | Division of Intramural Research (DIR, NIAID)
- 101007873/Cordis (Community Research and Development Information Service)
- RIA2018CO-2514/EC | H2020 | ERA-LEARN | European and Developing Countries Clinical Trials Partnership (EDCTP)
- P30CA125123/National Cancer Center (NCC)
- P30 ES030285/ES/NIEHS NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- R01 AI165721/AI/NIAID NIH HHS/United States
- P50MD015496/HHS | NIH | National Institute on Minority Health and Health Disparities (NIMHD)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

