Identification of Immune-Related Genes as Potential Biomarkers in Early Septic Shock
- PMID: 39348809
- PMCID: PMC11887992
- DOI: 10.1159/000540949
Identification of Immune-Related Genes as Potential Biomarkers in Early Septic Shock
Abstract
Introduction: Septic shock, a severe manifestation of infection-induced systemic immune response, poses a critical threat resulting in life-threatening multi-organ failure. Early diagnosis and intervention are imperative due to the potential for irreversible organ damage. However, specific and sensitive detection tools for the diagnosis of septic shock are still lacking.
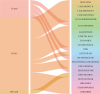
Methods: Gene expression files of early septic shock were obtained from the Gene Expression Omnibus (GEO) database. CIBERSORT analysis was used to evaluate immune cell infiltration. Genes related to immunity and disease progression were identified using weighted gene co-expression network analysis (WGCNA), followed by enrichment analysis. CytoHubba was then employed to identify hub genes, and their relationships with immune cells were explored through correlation analysis. Blood samples from healthy controls and patients with early septic shock were collected to validate the expression of hub genes, and an external dataset was used to validate their diagnostic efficacy.
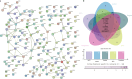
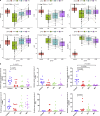
Results: Twelve immune cells showed significant infiltration differences in early septic shock compared to control, such as neutrophils, M0 macrophages, and natural killer cells. The identified immune and disease-related genes were mainly enriched in immune, cell signaling, and metabolism pathways. In addition, six hub genes were identified (PECAM1, F11R, ITGAL, ICAM3, HK3, and MCEMP1), all significantly associated with M0 macrophages and exhibiting an area under curve of over 0.7. These genes exhibited abnormal expression in patients with early septic shock. External datasets and real-time qPCR validation supported the robustness of these findings.
Conclusion: Six immune-related hub genes may be potential biomarkers for early septic shock.
Introduction: Septic shock, a severe manifestation of infection-induced systemic immune response, poses a critical threat resulting in life-threatening multi-organ failure. Early diagnosis and intervention are imperative due to the potential for irreversible organ damage. However, specific and sensitive detection tools for the diagnosis of septic shock are still lacking.
Methods: Gene expression files of early septic shock were obtained from the Gene Expression Omnibus (GEO) database. CIBERSORT analysis was used to evaluate immune cell infiltration. Genes related to immunity and disease progression were identified using weighted gene co-expression network analysis (WGCNA), followed by enrichment analysis. CytoHubba was then employed to identify hub genes, and their relationships with immune cells were explored through correlation analysis. Blood samples from healthy controls and patients with early septic shock were collected to validate the expression of hub genes, and an external dataset was used to validate their diagnostic efficacy.
Results: Twelve immune cells showed significant infiltration differences in early septic shock compared to control, such as neutrophils, M0 macrophages, and natural killer cells. The identified immune and disease-related genes were mainly enriched in immune, cell signaling, and metabolism pathways. In addition, six hub genes were identified (PECAM1, F11R, ITGAL, ICAM3, HK3, and MCEMP1), all significantly associated with M0 macrophages and exhibiting an area under curve of over 0.7. These genes exhibited abnormal expression in patients with early septic shock. External datasets and real-time qPCR validation supported the robustness of these findings.
Conclusion: Six immune-related hub genes may be potential biomarkers for early septic shock.
Keywords: Biomarkers; Immune infiltration; Septic shock; Weighted gene co-expression network analysis.
© 2024 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
The authors declare no conflicts of interest in this work.
Figures






References
-
- Font MD, Thyagarajan B, Khanna AK. Sepsis and septic shock: basics of diagnosis, pathophysiology and clinical decision making. Med Clin North Am. 2020;104(4):573–85. - PubMed
-
- Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365(9453):63–78. - PubMed
-
- Jacobi J. The pathophysiology of sepsis: 2021 update – part 2, organ dysfunction and assessment. Am J Health Syst Pharm. 2022;79(6):424–36. - PubMed
-
- Norse AB, Guirgis F, Black LP, DeVos EL. Updates and controversies in the early management of sepsis and septic shock. Emerg Med Pract. 2021;23(Suppl 4–2):1–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

