Proinflammatory immune cells disrupt angiogenesis and promote germinal matrix hemorrhage in prenatal human brain
- PMID: 39349662
- PMCID: PMC11537974
- DOI: 10.1038/s41593-024-01769-2
Proinflammatory immune cells disrupt angiogenesis and promote germinal matrix hemorrhage in prenatal human brain
Abstract
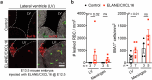
Germinal matrix hemorrhage (GMH) is a devastating neurodevelopmental condition affecting preterm infants, but why blood vessels in this brain region are vulnerable to rupture remains unknown. Here we show that microglia in prenatal mouse and human brain interact with nascent vasculature in an age-dependent manner and that ablation of these cells in mice reduces angiogenesis in the ganglionic eminences, which correspond to the human germinal matrix. Consistent with these findings, single-cell transcriptomics and flow cytometry show that distinct subsets of CD45+ cells from control preterm infants employ diverse signaling mechanisms to promote vascular network formation. In contrast, CD45+ cells from infants with GMH harbor activated neutrophils and monocytes that produce proinflammatory factors, including azurocidin 1, elastase and CXCL16, to disrupt vascular integrity and cause hemorrhage in ganglionic eminences. These results underscore the brain's innate immune cells in region-specific angiogenesis and how aberrant activation of these immune cells promotes GMH in preterm infants.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

















References
-
- Ma, T. et al. Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci.16, 1588–1597 (2013). - PubMed
MeSH terms
Grants and funding
- R01 NS128021/NS/NINDS NIH HHS/United States
- I01 BX001108/BX/BLRD VA/United States
- P01 NS083513/NS/NINDS NIH HHS/United States
- P50 HD105351/HD/NICHD NIH HHS/United States
- R01 NS132595/NS/NINDS NIH HHS/United States
- R01 NS124637/NS/NINDS NIH HHS/United States
- S10 OD028611/OD/NIH HHS/United States
- K08 NS116161/NS/NINDS NIH HHS/United States
- K12 HD000850/HD/NICHD NIH HHS/United States
- T32 GM144273/GM/NIGMS NIH HHS/United States
- R21 AG072232/AG/NIA NIH HHS/United States
- R01 NS129823/NS/NINDS NIH HHS/United States
- Grant #23IPA1051611/2023/American Heart Association (American Heart Association, Inc.)
- K99 MH129983/MH/NIMH NIH HHS/United States
- RF1 DA048790/DA/NIDA NIH HHS/United States
- R35 GM150987/GM/NIGMS NIH HHS/United States
- R00 MH129983/MH/NIMH NIH HHS/United States
- R21 NS133891/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

