Multivariate genomic analysis of 5 million people elucidates the genetic architecture of shared components of the metabolic syndrome
- PMID: 39349817
- PMCID: PMC11549047
- DOI: 10.1038/s41588-024-01933-1
Multivariate genomic analysis of 5 million people elucidates the genetic architecture of shared components of the metabolic syndrome
Abstract
Metabolic syndrome (MetS) is a complex hereditary condition comprising various metabolic traits as risk factors. Although the genetics of individual MetS components have been investigated actively through large-scale genome-wide association studies, the conjoint genetic architecture has not been fully elucidated. Here, we performed the largest multivariate genome-wide association study of MetS in Europe (nobserved = 4,947,860) by leveraging genetic correlation between MetS components. We identified 1,307 genetic loci associated with MetS that were enriched primarily in brain tissues. Using transcriptomic data, we identified 11 genes associated strongly with MetS. Our phenome-wide association and Mendelian randomization analyses highlighted associations of MetS with diverse diseases beyond cardiometabolic diseases. Polygenic risk score analysis demonstrated better discrimination of MetS and predictive power in European and East Asian populations. Altogether, our findings will guide future studies aimed at elucidating the genetic architecture of MetS.
© 2024. The Author(s).
Conflict of interest statement
A.C.F. is cofounder of Goodpath. P.T.E. reports personal fees from Bayer AG, Novartis and MyoKardia. P.N. reports personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co., Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, Merck and Novartis; scientific advisory board membership at Esperion Therapeutics, Preciseli and TenSixteen Bio; scientific cofounder status at TenSixteen Bio; equity in MyOme, Preciseli and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. W.-Y.P. was employed by the commercial company GENINUS. A.T. declares he is a cofounder and equity shareholder of GeneXwell Inc. A.T. is an advisor to InsideTracker. The other authors declare no competing interests.
Figures
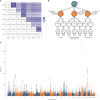
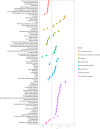
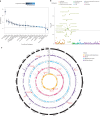
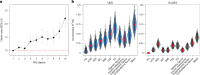

References
-
- Alberti, K. G. et al. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; National Heart, Lung, And Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation120, 1640–1645 (2009). - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

