Low protein diet protects the liver from Salmonella Typhimurium-mediated injury by modulating the mTOR/autophagy axis in macrophages
- PMID: 39349819
- PMCID: PMC11444042
- DOI: 10.1038/s42003-024-06932-w
Low protein diet protects the liver from Salmonella Typhimurium-mediated injury by modulating the mTOR/autophagy axis in macrophages
Abstract
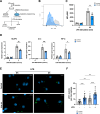
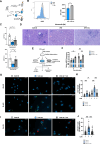
Western diets are the underlying cause of metabolic and liver diseases. Recent trend to limit the consumption of protein-rich animal products has become more prominent. This dietary change entails decreased protein consumption; however, it is still unknown how this affects innate immunity. Here, we studied the influence of a low protein diet (LPD) on the liver response to bacterial infection in mice. We found that LPD protects from Salmonella enterica serovar Typhimurium (S. Typhimurium)-induced liver damage. Bulk and single-cell RNA sequencing of murine liver cells showed reduced inflammation and upregulation of autophagy-related genes in myeloid cells in mice fed with LPD after S. Typhimurium infection. Mechanistically, we found reduced activation of the mammalian target of rapamycin (mTOR) pathway, whilst increased phagocytosis and activation of autophagy in LPD-programmed macrophages. We confirmed these observations in phagocytosis and mTOR activation in metabolically programmed human peripheral blood monocyte-derived macrophages. Together, our results support the causal role of dietary components on the fitness of the immune system.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. Robert A. Kingsley is an Editorial Board Member for Communications Biology, but was not involved in the editorial review of, nor the decision to publish this article.
Figures



References
-
- Cildir, G., Akincilar, S. C. & Tergaonkar, V. Chronic adipose tissue inflammation: all immune cells on the stage. Trends Mol. Med.19, 487–500 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- BB/P022073/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BBS/E/F/000PR10355/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BBS/E/F/000PR13632/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/CCG1860/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BBX011070/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/T008717/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/X011011/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- MR/T02934X/1/RCUK | Medical Research Council (MRC)
- BB/W002450/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/X011054/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- WT_/Wellcome Trust/United Kingdom
- BB/R012490/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/CCG1720/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- 220534/Z/20/Z/Wellcome Trust (Wellcome)
- 213731/Z/18/Z/Wellcome Trust (Wellcome)
- BBS/E/F/000PR13634/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
LinkOut - more resources
Full Text Sources
Miscellaneous

