Renal fibroblasts are involved in fibrogenic changes in kidney fibrosis associated with dysfunctional telomeres
- PMID: 39349834
- PMCID: PMC11541748
- DOI: 10.1038/s12276-024-01318-8
Renal fibroblasts are involved in fibrogenic changes in kidney fibrosis associated with dysfunctional telomeres
Erratum in
-
Author Correction: Renal fibroblasts are involved in fibrogenic changes in kidney fibrosis associated with dysfunctional telomeres.Exp Mol Med. 2024 Dec;56(12):2763. doi: 10.1038/s12276-024-01370-4. Exp Mol Med. 2024. PMID: 39633034 Free PMC article. No abstract available.
Abstract
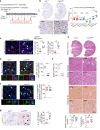
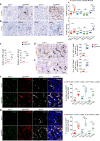
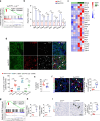
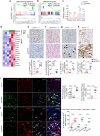
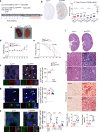
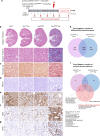
Tubulointerstitial fibrosis associated with chronic kidney disease (CKD) represents a global health care problem. We previously reported that short and dysfunctional telomeres lead to interstitial renal fibrosis; however, the cell-of-origin of kidney fibrosis associated with telomere dysfunction is currently unknown. We induced telomere dysfunction by deleting the Trf1 gene encoding a telomere-binding factor specifically in renal fibroblasts in both short-term and long-term life-long experiments in mice to identify the role of fibroblasts in renal fibrosis. Short-term Trf1 deletion in renal fibroblasts was not sufficient to trigger kidney fibrosis but was sufficient to induce inflammatory responses, ECM deposition, cell cycle arrest, fibrogenesis, and vascular rarefaction. However, long-term persistent deletion of Trf1 in fibroblasts resulted in kidney fibrosis accompanied by an elevated urinary albumin-to-creatinine ratio (uACR) and a decrease in mouse survival. These cellular responses lead to the macrophage-to-myofibroblast transition (MMT), endothelial-to-mesenchymal transition (EndMT), and partial epithelial-to-mesenchymal transition (EMT), ultimately causing kidney fibrosis at the humane endpoint (HEP) when the deletion of Trf1 in fibroblasts is maintained throughout the lifespan of mice. Our findings contribute to a better understanding of the role of dysfunctional telomeres in the onset of the profibrotic alterations that lead to kidney fibrosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

