Performance of somatic structural variant calling in lung cancer using Oxford Nanopore sequencing technology
- PMID: 39350042
- PMCID: PMC11441263
- DOI: 10.1186/s12864-024-10792-3
Performance of somatic structural variant calling in lung cancer using Oxford Nanopore sequencing technology
Abstract
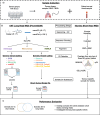
Background: Lung cancer is a heterogeneous disease and the primary cause of cancer-related mortality worldwide. Somatic mutations, including large structural variants, are important biomarkers in lung cancer for selecting targeted therapy. Genomic studies in lung cancer have been conducted using short-read sequencing. Emerging long-read sequencing technologies are a promising alternative to study somatic structural variants, however there is no current consensus on how to process data and call somatic events. In this study, we preformed whole genome sequencing of lung cancer and matched non-tumour samples using long and short read sequencing to comprehensively benchmark three sequence aligners and seven structural variant callers comprised of generic callers (SVIM, Sniffles2, DELLY in generic mode and cuteSV) and somatic callers (Severus, SAVANA, nanomonsv and DELLY in somatic modes).
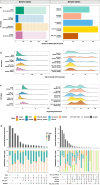
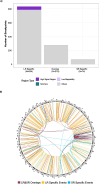
Results: Different combinations of aligners and variant callers influenced somatic structural variant detection. The choice of caller had a significant influence on somatic structural variant detection in terms of variant type, size, sensitivity, and accuracy. The performance of each variant caller was assessed by comparing to somatic structural variants identified by short-read sequencing. When compared to somatic structural variants detected with short-read sequencing, more events were detected with long-read sequencing. The mean recall of somatic variant events identified by long-read sequencing was higher for the somatic callers (72%) than generic callers (53%). Among the somatic callers when using the minimap2 aligner, SAVANA and Severus achieved the highest recall at 79.5% and 79.25% respectively, followed by nanomonsv with a recall of 72.5%.
Conclusion: Long-read sequencing can identify somatic structural variants in clincal samples. The longer reads have the potential to improve our understanding of cancer development and inform personalized cancer treatment.
Keywords: Benchmarking long read approaches; Long read sequencing; Small cell lung cancer; Somatic structural variants detection.
© 2024. The Author(s).
Conflict of interest statement
John V. Pearson and Nicola Waddell are co-founders of genomiQa. LL and NW were funded by Oxford Nanopore to present work from this study at meetings. The remaining authors declare that there are no competing interests.
Figures

References
-
- WHO. Cancer World Health Organization (Fact sheets). 2022. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer. Cited 2023 23rd Feb.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

