Determination of the frequency and distribution of APC, PIK3CA, and SMAD4 gene mutations in Ugandan patients with colorectal cancer
- PMID: 39350061
- PMCID: PMC11440721
- DOI: 10.1186/s12885-024-12967-3
Determination of the frequency and distribution of APC, PIK3CA, and SMAD4 gene mutations in Ugandan patients with colorectal cancer
Abstract
Uganda is a developing low-income country with a low incidence of colorectal cancer, which is steadily increasing. Ugandan colorectal cancer (CRC) patients are young and present with advanced-stage disease. In our population, there is a scarcity of genetic oncological studies, therefore, we investigated the mutational status of CRC tissues, focusing in particular on the adenomatous polyposis coli (APC), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and SMAD4 genes. Our objective was to determine whether there were any differences between other populations and Ugandan patients. We performed next-generation sequencing on the extracted DNA from formalin-fixed paraffin-embedded adenocarcinoma samples from 127 patients (mean (SD) age: 54.9 (16.0) years; male:female sex ratio: 1.2:1). Most tumours were located in the rectum 56 (44.1%), 14 (11%) tumours were high grade, and 96 (75.6%) were moderate grade CRC. Stage III + IV CRC tumours were found in 109 (85.8%) patients. We identified 48 variants of APC, including 9 novel APC mutations that were all pathogenic or deleterious. For PIK3CA, we found 19 variants, of which 9 were deleterious or pathogenic. Four PIK3CA novel pathogenic or deleterious variants were included (c.1397C > G, c.2399_2400insA, c.2621G > C, c.2632C > G). Three SMAD4 variants were reported, including two pathogenic or deleterious variants (c.1268G > T, c.556dupC) and one tolerant (c.563A > C) variant. One novel SMAD4 deleterious mutation (c.1268G > T) was reported. In conclusion, we provide clinicopathological information and new genetic variation data pertinent to CRC in Uganda.
Keywords: APC; Africa; Colorectal cancer; Genetics; Mutation; PIK3CA; SMAD4; Uganda; Variants.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

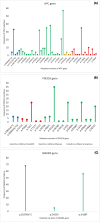
 , Missense
, Missense
 , Synonymous
, Synonymous
 , and Stop gain
, and Stop gain
 . A APC gene variants, B PIK3CA gene variants and C SMAD4 gene variants
. A APC gene variants, B PIK3CA gene variants and C SMAD4 gene variantsReferences
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

