OncomiR-181a promotes carcinogenesis by repressing the extracellular matrix proteoglycan decorin in hepatocellular carcinoma
- PMID: 39350070
- PMCID: PMC11443891
- DOI: 10.1186/s12876-024-03413-6
OncomiR-181a promotes carcinogenesis by repressing the extracellular matrix proteoglycan decorin in hepatocellular carcinoma
Abstract
Background: Proteoglycans are important tumor microenvironment extracellular matrix components. The regulation of key proteoglycans, such as decorin (DCN), by miRNAs has drawn attention since they have surfaced as novel therapeutic targets in cancer. Accordingly, this study aimed at identifying the impact of miR-181a in liver cancer and its regulatory role on the extracellular matrix proteoglycan, DCN, and hence on downstream oncogenes and tumor suppressor genes.
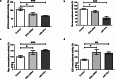
Results: DCN was under-expressed in 22 cirrhotic and HCC liver tissues compared to that in 11 healthy tissues of liver transplantation donors. Conversely, miR-181a was over-expressed in HCC liver tissues compared to that in healthy liver tissues. In silico analysis predicted that DCN 3'UTR harbors two high-score oncomiR-181a binding regions. This was validated by pmiRGLO luciferase reporter assay. Ectopic miR-181a expression into HuH-7 cells repressed the transcript and protein levels of DCN as assessed fluorometrically and by western blotting. DCN siRNAs showed similar results to miR-181a, where they both enhanced the cellular viability, proliferation, and clonogenicity. They also increased Myc and E2F and decreased p53 and Rb signaling as assessed using reporter vectors harboring p53, Rb, Myc, and E2F response elements. Our findings demonstrated that miR-181a directly downregulated the expression of its direct downstream target DCN, which in turn affected downstream targets related to cellular proliferation and apoptosis.
Conclusion: To our knowledge, this is the first study to unveil the direct targeting of DCN by oncomiR-181a. We also highlighted that miR-181a affects targets related to cellular proliferation in HCC which may be partly mediated through inhibition of DCN transcription. Thus, miR-181a could be a promising biomarker for the early detection and monitoring of liver cancer progression. This would pave the way for the future targeting of the oncomiR-181a as a therapeutic approach in liver cancer, where miR-181a-based therapy approach could be potentially combined with chemotherapy and immunotherapy for the management of liver cancer.
Keywords: Decorin (DCN); Hepatocellular carcinoma (HCC); Liver cancer; microRNA-181a (miR-181a).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


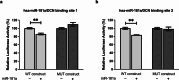

References
-
- Sweed D, Sweed E, Moaz I, Mosbeh A, Fayed Y, Elhamed SMA, Sweed E, Macshut M, Abdelsattar S, Kilany S, et al. The clinicopathological and prognostic factors of hepatocellular carcinoma: a 10-year tertiary center experience in Egypt. World J Surg Oncol. 2022;20. 10.1186/S12957-022-02764-2 - PMC - PubMed
MeSH terms
Substances
Grants and funding
- YRG-33464/Egyptian Science, Technology & Innovation Funding Authority
- YRG-33464/Egyptian Science, Technology & Innovation Funding Authority
- BARG-37096/Egyptian Science, Technology & Innovation Funding Authority
- BARG-37096/Egyptian Science, Technology & Innovation Funding Authority
- 412294938/Deutsche Forschungsgemeinschaft (DFG)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

