Multi-center external validation of an automated method segmenting and differentiating atypical lipomatous tumors from lipomas using radiomics and deep-learning on MRI
- PMID: 39351025
- PMCID: PMC11440245
- DOI: 10.1016/j.eclinm.2024.102802
Multi-center external validation of an automated method segmenting and differentiating atypical lipomatous tumors from lipomas using radiomics and deep-learning on MRI
Abstract
Background: As differentiating between lipomas and atypical lipomatous tumors (ALTs) based on imaging is challenging and requires biopsies, radiomics has been proposed to aid the diagnosis. This study aimed to externally and prospectively validate a radiomics model differentiating between lipomas and ALTs on MRI in three large, multi-center cohorts, and extend it with automatic and minimally interactive segmentation methods to increase clinical feasibility.
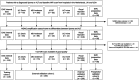
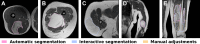
Methods: Three study cohorts were formed, two for external validation containing data from medical centers in the United States (US) collected from 2008 until 2018 and the United Kingdom (UK) collected from 2011 until 2017, and one for prospective validation consisting of data collected from 2020 until 2021 in the Netherlands. Patient characteristics, MDM2 amplification status, and MRI scans were collected. An automatic segmentation method was developed to segment all tumors on T1-weighted MRI scans of the validation cohorts. Segmentations were subsequently quality scored. In case of insufficient quality, an interactive segmentation method was used. Radiomics performance was evaluated for all cohorts and compared to two radiologists.
Findings: The validation cohorts included 150 (54% ALT), 208 (37% ALT), and 86 patients (28% ALT) from the US, UK and NL. Of the 444 cases, 78% were automatically segmented. For 22%, interactive segmentation was necessary due to insufficient quality, with only 3% of all patients requiring manual adjustment. External validation resulted in an AUC of 0.74 (95% CI: 0.66, 0.82) in US data and 0.86 (0.80, 0.92) in UK data. Prospective validation resulted in an AUC of 0.89 (0.83, 0.96). The radiomics model performed similar to the two radiologists (US: 0.79 and 0.76, UK: 0.86 and 0.86, NL: 0.82 and 0.85).
Interpretation: The radiomics model extended with automatic and minimally interactive segmentation methods accurately differentiated between lipomas and ALTs in two large, multi-center external cohorts, and in prospective validation, performing similar to expert radiologists, possibly limiting the need for invasive diagnostics.
Funding: Hanarth fonds.
Keywords: Deep learning-based segmentation; External validation; Lipomatous tumors; Magnetic resonance imaging; Prospective validation; Radiomics.
© 2024 The Author(s).
Conflict of interest statement
JV received a grant to institution from Qure.ai/HealthHolland/Enlitic; consulting fees from Tegus; payment to institution for lectures from Roche; travel grant from Qure.ai; participation on a data safety monitoring board or advisory board from Contextflow, Noaber Foundation, and NLC Ventures; leadership or fiduciary role on the steering committee of the PINPOINT Project (payment to institution from AstraZeneca) and RSNA Common Data Elements Steering Committee (unpaid); phantom shares in Contextflow and Quibim; chair scientific committee EuSoMII (unpaid); chair ESR value-based radiology subcommittee (unpaid); member editorial board European Journal of Radiology (unpaid). SD and RJ received support from the National Institute for Health Research (NIHR) Biomedical Research Centre at The Royal Marsden NHS Foundation Trust and The Institute of Cancer Research, London, and by The Royal Marsden Cancer Charity. RJ recieved consulting fees from Adaptimmune/Astex/Athenex/Bayer/B.I./Blueprint/Clinigen/Eisai/Epizyme/Daiichi/Deciphera/Immune Design/Immunicum/Karma Oncology/Lilly/Merck/PharmaMar/Tracon; LN is supported by the In Vivo Translational Imaging Shared Resources with funds from NCI P30CA093373; is principal investigator of a service agreement with United Imaging Healthcare; has been the PI of more than 1 service agreements with United Imaging Healthcare; is site PI of clinical trials supported by Novartis Pharmaceuticals Corporation; is PI of a clinical trial supported by Telix Pharmaceuticals; is PI of clinical trial supported by Lantheus Medical Imaging; is PI of a clinical trials supported by GE Healthcare; is Co-I of a clinical trial supported by Lilly; has a speaker engagement agreement with Lilly; served a panel reviewer for the European Health and Digital Executive Agency. UC Davis has a revenue-sharing agreement with United Imaging Healthcare that is based on uEXPLORER sales. SK is scientific director of the ICAI lab “Trustworthy AI for MRI”, a public-private research program partially funded by General Electric Healthcare. SK and MS received an unrestricted research grant from Stichting Hanarth Fonds, The Netherlands. MS acknowledge funding from the research project EuCanImage (European Union's Horizon 2020 research and innovation programme under grant agreement Nr. 95210). The other authors do not have any conflicts of interest.
Figures


References
-
- Johnson C.N., Ha A.S., Chen E., Davidson D. Lipomatous soft-tissue tumors. J Am Acad Orthop Surg. 2018;26(22):779–788. - PubMed
-
- Kooby D.A., Antonescu C.R., Brennan M., et al. Atypical lipomatous tumor/well-differentiated liposarcoma of the extremity and trunk wall: importance of histological subtype with treatment recommendations. Ann Surg Oncol. 2004;11:78–84. - PubMed
-
- Hogg M.E., Wayne J.D. Atypical lipomatous tumor/well-differentiated liposarcoma: what is it? Surg Oncol Clin. 2012;21(2):333–340. - PubMed
-
- Weiss S.W., Rao V.K. Well-differentiated liposarcoma (atypical lipoma) of deep soft tissue of the extremities, retroperitoneum, and miscellaneous sites. A follow-up study of 92 cases with analysis of the incidence of "dedifferentiation". Am J Surg Pathol. 1992;16(11):1051–1058. - PubMed
-
- Kransdorf M.J., Bancroft L.W., Peterson J.J., Murphey M.D., Foster W.C., Temple H.T. Imaging of fatty tumors: distinction of lipoma and well-differentiated liposarcoma. Radiology. 2002;224(1):99–104. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

