SARS-CoV-2 ferritin nanoparticle vaccines produce hyperimmune equine sera with broad sarbecovirus activity
- PMID: 39351195
- PMCID: PMC11440237
- DOI: 10.1016/j.isci.2024.110624
SARS-CoV-2 ferritin nanoparticle vaccines produce hyperimmune equine sera with broad sarbecovirus activity
Abstract
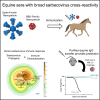
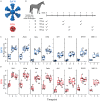
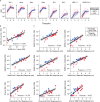
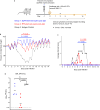
The rapid emergence of SARS-CoV-2 variants of concern (VoC) and the threat of future zoonotic sarbecovirus spillover emphasizes the need for broadly protective next-generation vaccines and therapeutics. We utilized SARS-CoV-2 spike ferritin nanoparticle (SpFN), and SARS-CoV-2 receptor binding domain ferritin nanoparticle (RFN) immunogens, in an equine model to elicit hyperimmune sera and evaluated its sarbecovirus neutralization and protection capacity. Immunized animals rapidly elicited sera with the potent neutralization of SARS-CoV-2 VoC, and SARS-CoV-1 pseudoviruses, and potent binding against receptor binding domains from sarbecovirus clades 1b, 1a, 2, 3, and 4. Purified equine polyclonal IgG provided protection against Omicron XBB.1.5 virus in the K18-hACE2 transgenic mouse model. These results suggest that SARS-CoV-2-based nanoparticle vaccines can rapidly produce a broad and protective sarbecovirus response in the equine model and that equine serum has therapeutic potential against emerging SARS-CoV-2 VoC and diverse sarbecoviruses, presenting a possible alternative or supplement to monoclonal antibody immunotherapies.
Keywords: Immunology; Virology.
© 2024 The Author(s).
Conflict of interest statement
W.H.C, A.H., P.V.T., J.L.J., K.M. and M.G.J. are named inventors on provisional patents describing SpFN molecules. S.J.K., V.D., N.L.M., and K.M. are named inventors on provisional patents describing monoclonal antibodies against coronaviruses. M.G.J. is named as an inventor on international patent application WO/2018/081318 and U.S. patents 10,960,070, and 11,964,010 entitled “Prefusion coronavirus spike proteins and their use.” K.M. is a current employee of Pfizer and may, therefore, be a shareholder. A.F., K.Mur., and J.Kau. are former or current employees of B.S.V. The other authors declare no competing interests.
Figures

References
-
- Racine T., Denizot M., Pannetier D., Nguyen L., Pasquier A., Raoul H., Saluzzo J.F., Kobinger G., Veas F., Herbreteau C.H. In Vitro Characterization and In Vivo Effectiveness of Ebola Virus Specific Equine Polyclonal F(ab')2. J. Infect. Dis. 2019;220:41–45. doi: 10.1093/infdis/jiz068. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

