Unveiling superior phenol detoxification and degradation ability in Candida tropicalis SHC-03: a comparative study with Saccharomyces cerevisiae BY4742
- PMID: 39351301
- PMCID: PMC11441332
- DOI: 10.3389/fmicb.2024.1442235
Unveiling superior phenol detoxification and degradation ability in Candida tropicalis SHC-03: a comparative study with Saccharomyces cerevisiae BY4742
Abstract
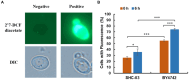
This study examined the phenol degradation capabilities and oxidative stress responses of Candida tropicalis SHC-03, demonstrating its metabolic superiority and resilience compared to Saccharomyces cerevisiae BY4742 in a culture medium with phenol as the sole carbon source. Through comparative growth, transcriptomic, and metabolomic analyses under different phenol concentrations, this study revealed C. tropicalis SHC-03's specialized adaptations for thriving in phenol as the sole carbon source environments. These include a strategic shift from carbohydrate metabolism to enhanced phenol degradation pathways, highlighted by the significant upregulation of genes for Phenol 2-monoxygenase and Catechol 1,2-dioxygenase. Despite phenol levels reaching 1.8 g/L, C. tropicalis exhibits a robust oxidative stress response, efficiently managing ROS through antioxidative pathways and the upregulation of genes for peroxisomal proteins like PEX2, PEX13, and PMP34. Concurrently, there was significant upregulation of genes associated with membrane components and transmembrane transporters, enhancing the cell's capacity for substance exchange and signal transduction. Especially, when the phenol concentration was 1.6 g/L and 1.8 g/L, the degradation rates of C. tropicalis towards it were 99.47 and 95.91%, respectively. Conversely, S. cerevisiae BY4742 shows limited metabolic response, with pronounced growth inhibition and lack of phenol degradation. Therefore, our study not only sheds light on the molecular mechanisms underpinning phenol tolerance and degradation in C. tropicalis but also positions this yeast as a promising candidate for environmental and industrial processes aimed at mitigating phenol pollution.
Keywords: Candida tropicalis; Saccharomyces cerevisiae; degradation; phenol; reactive oxygen species.
Copyright © 2024 Li, Chen, Tang, Shu, Zhang, Tang, Li, Jiang, Shen, Yang, Wang and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


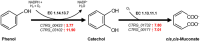
Similar articles
-
Isolation and partial characterization of catechol 1,2-dioxygenase of phenol degrading yeast Candida tropicalis.Neuro Endocrinol Lett. 2009;30 Suppl 1:80-7. Neuro Endocrinol Lett. 2009. PMID: 20027149
-
Isolation of cytoplasmic NADPH-dependent phenol hydroxylase and catechol-1,2-dioxygenase from Candida tropicalis yeast.Interdiscip Toxicol. 2008 Dec;1(3-4):225-30. doi: 10.2478/v10102-010-0046-7. Interdiscip Toxicol. 2008. PMID: 21218120 Free PMC article.
-
Biodegradation of phenol by a halotolerant versatile yeast Candida tropicalis SDP-1 in wastewater and soil under high salinity conditions.J Environ Manage. 2021 Jul 1;289:112525. doi: 10.1016/j.jenvman.2021.112525. Epub 2021 Apr 6. J Environ Manage. 2021. PMID: 33836438
-
Isolation and characterization of Candida tropicalis B: a promising yeast strain for biodegradation of petroleum oil in marine environments.Microb Cell Fact. 2024 Jan 13;23(1):20. doi: 10.1186/s12934-023-02292-y. Microb Cell Fact. 2024. PMID: 38218907 Free PMC article.
-
Biodegradation of phenol and 4-chlorophenol by the yeast Candida tropicalis.Biodegradation. 2007 Dec;18(6):719-29. doi: 10.1007/s10532-007-9100-3. Epub 2007 Jan 24. Biodegradation. 2007. PMID: 17245562
Cited by
-
Gene cloning, phenol-responsive transcriptional profiling and recombinant protein characterization of phenol hydroxylase in Candida tropicalis GY8.Braz J Microbiol. 2025 Sep;56(3):1985-1995. doi: 10.1007/s42770-025-01685-y. Epub 2025 Jun 2. Braz J Microbiol. 2025. PMID: 40455391
References
-
- Adeboye T. P. (2016). Mapping phenolics metabolism and metabolic engineering of Saccharomyces cerevisiae for increased endogenous catabolism of phenolic compounds. Gothenburg: Chalmers University of Technology.
-
- Agarry S. E., Durojaiye A. O., Solomon B. O. (2008). Microbial degradation of phenols: a review. Int. J. Environ. Pollut. 32, 12–28. doi: 10.1504/IJEP.2008.016895 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

