Molecular mechanisms of emerging inflammasome complexes and their activation and signaling in inflammation and pyroptosis
- PMID: 39351983
- PMCID: PMC11742652
- DOI: 10.1111/imr.13406
Molecular mechanisms of emerging inflammasome complexes and their activation and signaling in inflammation and pyroptosis
Abstract
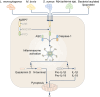
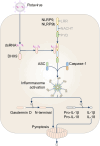
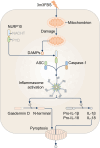
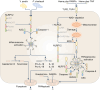
Inflammasomes are multi-protein complexes that assemble within the cytoplasm of mammalian cells in response to pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), driving the secretion of the pro-inflammatory cytokines IL-1β and IL-18, and pyroptosis. The best-characterized inflammasome complexes are the NLRP3, NAIP-NLRC4, NLRP1, AIM2, and Pyrin canonical caspase-1-containing inflammasomes, and the caspase-11 non-canonical inflammasome. Newer inflammasome sensor proteins have been identified, including NLRP6, NLRP7, NLRP9, NLRP10, NLRP11, NLRP12, CARD8, and MxA. These inflammasome sensors can sense PAMPs from bacteria, viruses and protozoa, or DAMPs in the form of mitochondrial damage, ROS, stress and heme. The mechanisms of action, physiological relevance, consequences in human diseases, and avenues for therapeutic intervention for these novel inflammasomes are beginning to be realized. Here, we discuss these emerging inflammasome complexes and their putative activation mechanisms, molecular and signaling pathways, and physiological roles in health and disease.
Keywords: PANoptosis; PANoptosome; autoimmunity; autoinflammation; bacteria; cancer; caspase‐1; caspase‐11; caspase‐4; caspase‐5; cell death; cytokines; gasdermin D; immunity; infection; inflammatory caspases; interferons; lipopolysaccharide; parasites; pattern‐recognition receptors; viruses.
© 2024 The Author(s). Immunological Reviews published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1‐13. - PubMed
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805‐820. - PubMed
-
- Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660‐665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

