Interpretable machine learning uncovers epithelial transcriptional rewiring and a role for Gelsolin in COPD
- PMID: 39352744
- PMCID: PMC11601586
- DOI: 10.1172/jci.insight.180239
Interpretable machine learning uncovers epithelial transcriptional rewiring and a role for Gelsolin in COPD
Abstract
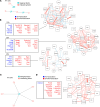
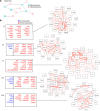
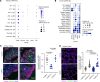
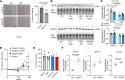
Transcriptomic analyses have advanced the understanding of complex disease pathophysiology including chronic obstructive pulmonary disease (COPD). However, identifying relevant biologic causative factors has been limited by the integration of high dimensionality data. COPD is characterized by lung destruction and inflammation, with smoke exposure being a major risk factor. To define previously unknown biological mechanisms in COPD, we utilized unsupervised and supervised interpretable machine learning analyses of single-cell RNA-Seq data from the mouse smoke-exposure model to identify significant latent factors (context-specific coexpression modules) impacting pathophysiology. The machine learning transcriptomic signatures coupled to protein networks uncovered a reduction in network complexity and new biological alterations in actin-associated gelsolin (GSN), which was transcriptionally linked to disease state. GSN was altered in airway epithelial cells in the mouse model and in human COPD. GSN was increased in plasma from patients with COPD, and smoke exposure resulted in enhanced GSN release from airway cells from patients with COPD. This method provides insights into rewiring of transcriptional networks that are associated with COPD pathogenesis and provides a translational analytical platform for other diseases.
Keywords: COPD; Cell biology; Cytoskeleton; Pulmonology.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

