RNA polymerase II transcription initiation in holo-TFIID-depleted mouse embryonic stem cells
- PMID: 39352809
- PMCID: PMC11551524
- DOI: 10.1016/j.celrep.2024.114791
RNA polymerase II transcription initiation in holo-TFIID-depleted mouse embryonic stem cells
Abstract
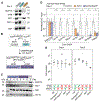
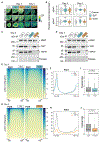
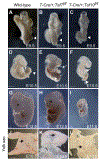
The recognition of core promoter sequences by TFIID is the first step in RNA polymerase II (Pol II) transcription initiation. Metazoan holo-TFIID is a trilobular complex, composed of the TATA binding protein (TBP) and 13 TBP-associated factors (TAFs). Why and how TAFs are necessary for the formation of TFIID domains and how they contribute to transcription initiation remain unclear. Inducible TAF7 or TAF10 depletion, followed by comprehensive analysis of TFIID subcomplex formation, chromatin binding, and nascent transcription in mouse embryonic stem cells, result in the formation of a TAF7-lacking TFIID or a minimal core-TFIID complex, respectively. These partial complexes support TBP recruitment at promoters and nascent Pol II transcription at most genes early after depletion, but importantly, TAF10 is necessary for efficient Pol II pausing. We show that partially assembled TFIID complexes can sustain Pol II transcription initiation but cannot replace holo-TFIID over several cell divisions and/or development.
Keywords: CP: Molecular biology; RNA polymerase II; TAF10; TAF7; TATA binding protein; TBP; TBP-associated factor; TFIID; complex assembly; mouse embryonic stem cells; nascent transcription.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

