METTL3-Mediated N 6 -Methyladenosine mRNA Modification and cGAS-STING Pathway Activity in Kidney Fibrosis
- PMID: 39352860
- PMCID: PMC11452136
- DOI: 10.1681/ASN.0000000000000428
METTL3-Mediated N 6 -Methyladenosine mRNA Modification and cGAS-STING Pathway Activity in Kidney Fibrosis
Abstract
Background: Chemical modifications on RNA profoundly affect RNA function and regulation. m6A, the most abundant RNA modification in eukaryotes, plays a pivotal role in diverse cellular processes and disease mechanisms. However, its importance is understudied in human CKD samples regarding its influence on pathological mechanisms.
Methods: Liquid chromatography–tandem mass spectrometry and methylated RNA immunoprecipitation sequencing were used to examine alterations in m6A levels and patterns in CKD samples. Overexpression of the m6A writer METTL3 in cultured kidney tubular cells was performed to confirm the effect of m6A in tubular cells and explore the biological functions of m6A modification on target genes. In addition, tubule-specific deletion of Mettl3 (Ksp-Cre Mettl3f/f) mice and antisense oligonucleotides inhibiting Mettl3 expression were used to reduce m6A modification in an animal kidney disease model.
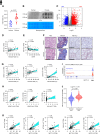
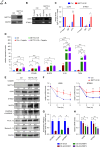
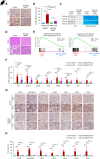
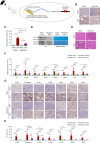
Results: By examining 127 human CKD samples, we observed a significant increase in m6A modification and METTL3 expression in diseased kidneys. Epitranscriptomic analysis unveiled an enrichment of m6A modifications in transcripts associated with the activation of inflammatory signaling pathways, particularly the cyclic guanosine monophosphate–AMP synthase (cGAS)-stimulator of IFN genes (STING) pathway. m6A hypermethylation increased mRNA stability in cGAS and STING1 as well as elevated the expression of key proteins within the cGAS-STING pathway. Both the tubule-specific deletion of Mettl3 and the use of antisense oligonucleotides to inhibit Mettl3 expression protected mice from inflammation, reduced cytokine expression, decreased immune cell recruitment, and attenuated kidney fibrosis.
Conclusions: Our research revealed heightened METTL3-mediated m6A modification in fibrotic kidneys, particularly enriching the cGAS-STING pathway. This hypermethylation increased mRNA stability for cGAS and STING1, leading to sterile inflammation and fibrosis.
Copyright © 2024 by the American Society of Nephrology
Conflict of interest statement
Disclosure forms, as provided by each author, are available with the online version of the article at
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

