PPP3CB overexpression mediates EGFR TKI resistance in lung tumors via calcineurin/MEK/ERK signaling
- PMID: 39353739
- PMCID: PMC11447527
- DOI: 10.26508/lsa.202402873
PPP3CB overexpression mediates EGFR TKI resistance in lung tumors via calcineurin/MEK/ERK signaling
Abstract
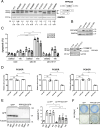
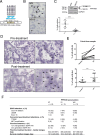
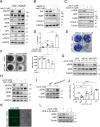
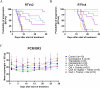
Despite initial high response rates to first-line EGFR TKI, all non-small-cell lung cancer (NSCLC) with EGFR-activating mutation will ultimately develop resistance to treatment. Identification of resistance mechanisms is critical to adapt treatment and improve patient outcomes. Here, we show that a PPP3CB transcript that encodes full-length catalytic subunit 2B of calcineurin accumulates in EGFR-mutant NSCLC cells with acquired resistance against different EGFR TKIs and in post-progression biopsies of NSCLC patients treated with EGFR TKIs. Neutralization of PPP3CB by siRNA or inactivation of calcineurin by cyclosporin A induces apoptosis in resistant cells treated with EGFR TKIs. Mechanistically, EGFR TKIs increase the cytosolic level of calcium and trigger activation of a calcineurin/MEK/ERK pathway that prevents apoptosis. Combining EGFR, calcineurin, and MEK inhibitors overcomes resistance to EGFR TKI in both in vitro and in vivo models. Our results identify PPP3CB overexpression as a new mechanism of acquired resistance to EGFR TKIs, and provide a promising therapeutic approach for NSCLC patients that progress under TKI treatment.
© 2024 Gazzeri et al.
Conflict of interest statement
N Girard has received research grants/support from AbbVie, Amgen, AstraZeneca, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi-Sankyo, Gilead, Hoffmann-La Roche, Janssen, Leo Pharma, Lilly, Merck Serono, Merck Sharp & Dohme, Novartis, Sanofi, and Sivan; has provided consultative services for AbbVie, Amgen, AstraZeneca, BeiGene, Bristol Myers Squibb, Daiichi-Sankyo, Gilead, Ipsen, Hoffmann-La Roche, Janssen, Leo Pharma, Lilly, Merck Sharp & Dohme, Mirati, Novartis, Pfizer, Pierre Fabre, Sanofi, and Takeda; has participated on a data safety monitoring board for Hoffmann-La Roche; and has a family member employed with AstraZeneca. A-C Toffart has received consulting fees, payment, or honoraria for lectures and presentations from Astra Zeneca, BMS, Janssen, MSD, Pfizer, Sanofi, Roche, and Takeda; and support for attending meetings and/or travel from Astra Zeneca, Pfizer, MSD, and Roche. G Berardi received support for attending meetings and/or travel from Pfizer.
Figures








References
-
- Dagogo-Jack I, Piotrowska Z, Cobb R, Banwait M, Lennerz JK, Hata AN, Digumarthy SR, Sequist LV (2019) Response to the combination of osimertinib and trametinib in a patient with egfr-mutant nsclc harboring an acquired braf fusion. J Thorac Oncol 14: e226–e228. 10.1016/j.jtho.2019.05.046 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
