Deciphering the impact of PROM1 alternative splicing on human photoreceptor development and maturation
- PMID: 39353897
- PMCID: PMC11445533
- DOI: 10.1038/s41419-024-07105-7
Deciphering the impact of PROM1 alternative splicing on human photoreceptor development and maturation
Abstract
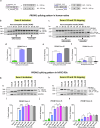
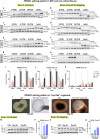
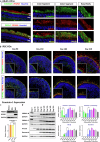
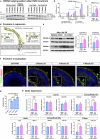
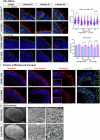
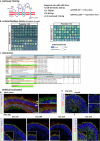
Alternative splicing (AS) is a crucial mechanism contributing to proteomic diversity, which is highly regulated in tissue- and development-specific patterns. Retinal tissue exhibits one of the highest levels of AS. In particular, photoreceptors have a distinctive AS pattern involving the inclusion of microexons not found in other cell types. PROM1 whose encoded protein Prominin-1 is located in photoreceptor outer segments (OSs), undergoes exon 4 inclusion from the 12th post-conception week of human development through adulthood. Exon 4 skipping in PROM1 is associated with late-onset mild maculopathy, however its role in photoreceptor maturation and function is unknown. In this study retinal organoids, a valuable model system, were employed in combination with phosphorodiamidate morpholino oligos (PMOs) to assess the role of exon 4 AS in the development of human retina. Retinal organoids were treated with the PMOs for four weeks after which RT-PCR, western blotting and immunofluorescence analysis were performed to assess exon 4 exclusion and its impact on photoreceptors. The transcriptome of treated ROs was studied by bulk RNA-Seq. Our data demonstrate that 55% skipping of PROM1 exon 4 resulted in decreased Prominin-1 expression by 40%, abnormal accumulation of cones in the basal side of the retinal organoids as well as detectable cone photoreceptor cilium defects. Transcriptomic and western blot analyses revealed decreased expression of cone, inner segment and connecting cilium basal body markers, increased expression of genes associated with stress response and the ubiquitin-proteasome system, and downregulation of autophagy. Importantly, the use of retinal organoids provides a valuable platform to study AS and unravel disease mechanisms in a more physiologically relevant context, opening avenues for further research and potential therapeutic interventions. Together our data indicate that cones may be more sensitive to PROM1 exon 4 skipping and/or reduced Prominin-1 expression, corroborating the pathogenesis of late-onset mild maculopathy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Weisschuh N, Buena-Atienza E, Wissinger B. Splicing mutations in inherited retinal diseases. Progr Retinal Eye Res. 2021;80:100874. - PubMed
-
- Corbeil D. Prominin-1 (CD133): New Insights on Stem & Cancer Stem Cell Biology. Advances in Experimental Medicine and Biology. New York: Springer Science+Business Media; 2013. 1–255 p. Available from: http://www.springer.com/series/5584.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

