GPR56 facilitates hepatocellular carcinoma metastasis by promoting the TGF-β signaling pathway
- PMID: 39353900
- PMCID: PMC11445230
- DOI: 10.1038/s41419-024-07095-6
GPR56 facilitates hepatocellular carcinoma metastasis by promoting the TGF-β signaling pathway
Abstract
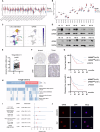
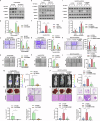
The metastasis of hepatocellular carcinoma (HCC) poses a significant threat to the survival of patients. G protein-coupled receptor 56 (GPR56) has garnered extensive attention within malignant tumor research and plays a crucial role in cellular surface signal transmission. Nonetheless, its precise function in HCC remains ambiguous. Our investigation reveals a notable rise in GPR56 expression levels in human HCC cases, with heightened GPR56 levels correlating with unfavorable prognoses. GPR56 regulates TGF-β pathway by interacting with TGFBR1, thereby promoting HCC metastasis. At the same time, GPR56 is subject to regulation by the canonical cascade of TGF-β signaling, thereby establishing a positive feedback loop. Furthermore, the combination application of TGFBR1 inhibitor galunisertib (GAL) and GPR56 inhibitor Dihydromunduletone (DHM), significantly inhibits HCC metastasis. Interventions towards this signaling pathway could offer a promising therapeutic approach to effectively impede the metastasis of GPR56-mediated HCC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

