Genomic evidence of two-staged transmission of the early seventh cholera pandemic
- PMID: 39353924
- PMCID: PMC11445481
- DOI: 10.1038/s41467-024-52800-w
Genomic evidence of two-staged transmission of the early seventh cholera pandemic
Abstract
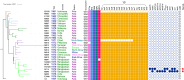
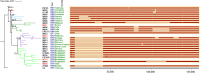
The seventh cholera pandemic started in 1961 in Indonesia and spread across the world in three waves in the decades that followed. Here, we utilised genomic evidence to detail the first wave of the seventh pandemic. Genomes of 22 seventh pandemic Vibrio cholerae isolates from 1961 to 1979 were completely sequenced. Together with 152 publicly available genomes from the same period, they fell into seven phylogenetic clusters (CL1-CL7). By multilevel genome typing (MGT), all were assigned to MGT2 ST1 (Wave 1) except three isolates in CL7 which were typed as MGT2 ST2 (Wave 2). The Wave 1 seventh pandemic expanded in two stages, with Stage 1 (CL1-CL5) spread across Asia and Stage 2 (CL6 and CL7) spread to the Middle East and Africa. Three non-synonymous mutations, one each, in three regulatory genes, csrD (global regulator), acfB (chemotaxis), and luxO (quorum sensing) may have critically contributed to its pandemicity. The three MGT2 ST2 isolates in CL7 were the progenitors of Wave 2 and evolved from within Wave 1 with acquisition of a novel IncA/C plasmid. Our findings provide new insight into the evolution and transmission of the early seventh pandemic, which may aid future cholera prevention and control.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Robert, S. Reports on the epidemic cholera which has raged throughout Hindostan and the peninsula of India, since August 1817. Published under the Authority Of Government. Med. Chir. J.2, 547–557 (1820).
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

