CHIP ameliorates nonalcoholic fatty liver disease via promoting K63- and K27-linked STX17 ubiquitination to facilitate autophagosome-lysosome fusion
- PMID: 39353976
- PMCID: PMC11445385
- DOI: 10.1038/s41467-024-53002-0
CHIP ameliorates nonalcoholic fatty liver disease via promoting K63- and K27-linked STX17 ubiquitination to facilitate autophagosome-lysosome fusion
Erratum in
-
Author Correction: CHIP ameliorates nonalcoholic fatty liver disease via promoting K63- and K27-linked STX17 ubiquitination to facilitate autophagosome-lysosome fusion.Nat Commun. 2024 Dec 4;15(1):10567. doi: 10.1038/s41467-024-55039-7. Nat Commun. 2024. PMID: 39632833 Free PMC article. No abstract available.
Abstract
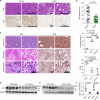
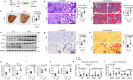
The fusion of autophagosomes and lysosomes is essential for the prevention of nonalcoholic fatty liver disease (NAFLD). Here, we generate a hepatocyte-specific CHIP knockout (H-KO) mouse model that develops NAFLD more rapidly in response to a high-fat diet (HFD) or high-fat, high-fructose diet (HFHFD). The accumulation of P62 and LC3 in the livers of H-KO mice and CHIP-depleted cells indicates the inhibition of autophagosome-lysosome fusion. AAV8-mediated overexpression of CHIP in the murine liver slows the progression of NAFLD induced by HFD or HFHFD feeding. Mechanistically, CHIP induced K63- and K27-linked polyubiquitination at the lysine 198 residue of STX17, resulting in increased STX17-SNAP29-VAMP8 complex formation. The STX17 K198R mutant was not ubiquitinated by CHIP; it interfered with its interaction with VAMP8, rendering STX17 incapable of inhibiting steatosis development in mice. These results indicate that a signaling regulatory mechanism involving CHIP-mediated non-degradative ubiquitination of STX17 is necessary for autophagosome-lysosome fusion.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Wai-Sun Wong, V., Ekstedt, M., Lai-Hung Wong, G. & Hagstrom, H. Changing epidemiology, global trends and implications for outcomes of NAFLD. J. Hepatol.79, 842–852 (2023). - PubMed
-
- Khan, R. S., Bril, F., Cusi, K. & Newsome, P. N. Modulation of insulin resistance in nonalcoholic fatty liver disease. Hepatology70, 711–724 (2019). - PubMed
-
- Rinella, M.E. et al. A multi-society Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol.78, 1966–1986 (2023). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

