The phyllosphere of Nigerian medicinal plants, Euphorbia lateriflora and Ficus thonningii is inhabited by a specific microbiota
- PMID: 39354019
- PMCID: PMC11448504
- DOI: 10.1038/s41598-024-68001-w
The phyllosphere of Nigerian medicinal plants, Euphorbia lateriflora and Ficus thonningii is inhabited by a specific microbiota
Abstract
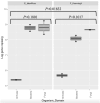
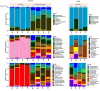
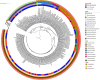
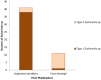
The microbiota of medicinal plants is known to be highly specific and can contribute to medicinal activity. However, the majority of plant species have not yet been studied. Here, we investigated the phyllosphere composition of two common Nigerian medicinal plants, Euphorbia lateriflora and Ficus thonningii, by a polyphasic approach combining analyses of metagenomic DNA and isolates. Microbial abundance estimated via qPCR using specific marker gene primers showed that all leaf samples were densely colonized, with up to 108 per gram of leaf, with higher bacterial and fungal abundance than Archaea. While no statistically significant differences between both plant species were found for abundance, amplicon sequencing of 16S rRNA and ITS genes revealed distinct microbiota compositions. Only seven of the 27 genera isolated were represented on both plants, e.g. dominant Sphingomonas spp., and numerous members of Xanthomonadaceae and Enterobacteriaceae. The most dominant fungal families on both plants were Cladosporiaceae, Mycosphaerellaceae and Trichosphaeriaceae. In addition, 225 plant-specific isolates were identified, with Pseudomonadota and Enterobacteriaceae being dominant. Interestingly, 29 isolates are likely species previously unknown, and 14 of these belong to Burkholderiales. However, a high proportion, 56% and 40% of the isolates from E. lateriflora and F. thonningii, respectively, were characterized as various Escherichia coli. The growth of most of the bacterial isolates was not influenced by extractable secondary metabolites of plants. Our results suggest that a specific and diverse microbial community inhabits the leaves of both E. lateriflora and F. thonningii, including potentially new species and producers of antimicrobials.
Keywords: Euphorbia lateriflora; Ficus thonningii; Antimicrobials; Medicinal plants; Phyllosphere.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Vorholt, J. A. Microbial life in the phyllosphere. Nat. Publ. Gr.10, 828–840 (2012). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

