Ladinin-1 in actin arcs of oral squamous cell carcinoma is involved in cell migration and epithelial phenotype
- PMID: 39354061
- PMCID: PMC11445451
- DOI: 10.1038/s41598-024-74041-z
Ladinin-1 in actin arcs of oral squamous cell carcinoma is involved in cell migration and epithelial phenotype
Abstract
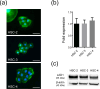
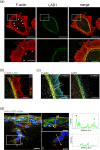
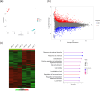
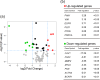
Histopathologically, oral squamous cell carcinoma (OSCC) consists of well-defined interfaces with adjacent non-cancerous epithelium. Previously, we found that SCC tissues expressed higher levels of specific proteins at this interface. Ladinin-1 (LAD1) is one of the specific molecules that has increased expressions in cancer fronts; however, its function in OSCC is unknown. Therefore, this study aimed to elucidate the function of LAD1 in human OSCC cells. LAD1 was localized on the actin arc at the distal periphery of cell clusters in the OSCC cell lines HSC-2, HSC-3, and HSC-4. When LAD1 was knocked down, cellular migration was repressed in wound scratch assays but was reversed in three-dimensional collagen gel invasion assays. Characteristic LAD1 localization along actin arcs forming the leading edge of migrating cells was diminished with loss of filopodia formation and ruffling in knockdown cells, in which the expression levels of cell motility-related genes-p21-activated kinase 1 (PAK1) and caveolin-1 (CAV1)-were upregulated and downregulated, respectively. LAD1 expression was also associated with the downregulation of vimentin and increased histological differentiation of OSCC. These results suggest that LAD1 is involved in actin dynamics during filopodia and lamellipodia formation, and in maintaining the epithelial phenotype of OSCC cells.
Keywords: Actin arc; Ladinin-1; Squamous cell carcinoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Abé, T. et al. Proteomic and histopathological characterization of the interface between oral squamous cell carcinoma invasion fronts and non-cancerous epithelia. Exp. Mol. Pathol. 102, 327–336 (2017). - PubMed
-
- Ishiko, A. et al. 97-kDa linear IgA bullous dermatosis (LAD) antigen localizes to the Lamina lucida of the epidermal basement membrane. J. Investig. Dermatol. 106, 739–743 (1996). - PubMed
-
- Marinkovich, M. P., Taylor, T. B., Keene, D. R., Burgeson, R. E. & Zone, J. J. LAD-1, the linear IgA bullous dermatosis autoantigen, is a novel 120-kDa anchoring filament protein synthesized by epidermal cells. J. Investig. Dermatol. 106, 734–738 (1996). - PubMed
-
- Roth, L. et al. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci. Signal. 11, eaan0949 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

