Derivation and validation of generalized sepsis-induced acute respiratory failure phenotypes among critically ill patients: a retrospective study
- PMID: 39354616
- PMCID: PMC11445942
- DOI: 10.1186/s13054-024-05061-4
Derivation and validation of generalized sepsis-induced acute respiratory failure phenotypes among critically ill patients: a retrospective study
Abstract
Background: Septic patients who develop acute respiratory failure (ARF) requiring mechanical ventilation represent a heterogenous subgroup of critically ill patients with widely variable clinical characteristics. Identifying distinct phenotypes of these patients may reveal insights about the broader heterogeneity in the clinical course of sepsis, considering multi-organ dynamics. We aimed to derive novel phenotypes of sepsis-induced ARF using observational clinical data and investigate the generalizability of the derived phenotypes.
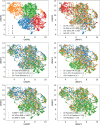
Methods: We performed a multi-center retrospective study of ICU patients with sepsis who required mechanical ventilation for ≥ 24 h. Data from two different high-volume academic hospital centers were used, where all phenotypes were derived in MICU of Hospital-I (N = 3225). The derived phenotypes were validated in MICU of Hospital-II (N = 848), SICU of Hospital-I (N = 1112), and SICU of Hospital-II (N = 465). Clinical data from 24 h preceding intubation was used to derive distinct phenotypes using an explainable machine learning-based clustering model interpreted by clinical experts.
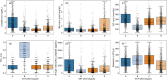
Results: Four distinct ARF phenotypes were identified: A (severe multi-organ dysfunction (MOD) with a high likelihood of kidney injury and heart failure), B (severe hypoxemic respiratory failure [median P/F = 123]), C (mild hypoxia [median P/F = 240]), and D (severe MOD with a high likelihood of hepatic injury, coagulopathy, and lactic acidosis). Patients in each phenotype showed differences in clinical course and mortality rates despite similarities in demographics and admission co-morbidities. The phenotypes were reproduced in external validation utilizing the MICU of Hospital-II and SICUs from Hospital-I and -II. Kaplan-Meier analysis showed significant difference in 28-day mortality across the phenotypes (p < 0.01) and consistent across MICU and SICU of both Hospital-I and -II. The phenotypes demonstrated differences in treatment effects associated with high positive end-expiratory pressure (PEEP) strategy.
Conclusion: The phenotypes demonstrated unique patterns of organ injury and differences in clinical outcomes, which may help inform future research and clinical trial design for tailored management strategies.
Keywords: Acute respiratory failure; Critical care; Phenotype; Sepsis-induced ARF; Unsupervised machine learning.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Derivation and Validation of Generalized Sepsis-induced Acute Respiratory Failure Phenotypes Among Critically Ill Patients: A Retrospective Study.Res Sq [Preprint]. 2024 Apr 30:rs.3.rs-4307475. doi: 10.21203/rs.3.rs-4307475/v1. Res Sq. 2024. Update in: Crit Care. 2024 Oct 1;28(1):321. doi: 10.1186/s13054-024-05061-4. PMID: 38746442 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

