HIV-1 inhibits IFITM3 expression to promote the infection of megakaryocytes
- PMID: 39354676
- PMCID: PMC11992561
- DOI: 10.1093/jmcb/mjae042
HIV-1 inhibits IFITM3 expression to promote the infection of megakaryocytes
Abstract
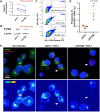
Despite an undetectable plasma viral load as a result of antiretroviral therapy, HIV-1-infected individuals with poor immune reconstitution harbor infectious HIV-1 within their platelets. Megakaryocytes, as platelet precursors, are the likely cellular origin of these HIV-1-containing platelets. To investigate the mechanisms that allow megakaryocytes to support HIV-1 infection, we established in vitro models of viral infection using hematopoietic stem cell-derived megakaryocytes and the megakaryocytic MEG-01 cell line. We observed HIV-1 DNA provirus integration into the megakaryocyte cell genome, self-limiting virus production, and HIV-1 protein and RNA compartmentalization, which are hallmarks of HIV-1 infection in myeloid cells. In addition, following HIV-1 infection of megakaryocyte precursors, the expression of interferon-induced transmembrane protein 3 (IFITM3), an antiviral factor constitutively expressed in megakaryocytes, was inhibited in terminally differentiated HIV-1-infected megakaryocytes. IFITM3 knockdown in MEG-01 cells prior to infection led to enhanced HIV-1 infection, indicating that IFITM3 acts as an HIV-1 restriction factor in megakaryocytes. Together, these findings indicate that megakaryocyte precursors are susceptible to HIV-1 infection, leading to terminally differentiated megakaryocytes harboring virus in a process regulated by IFITM3. Megakaryocytes may thus constitute a neglected HIV-1 reservoir that warrants further study in order to develop improved antiretroviral therapies and to facilitate HIV-1 eradication.
Keywords: HIV-1; IFITM3; megakaryocytes.
© The Author(s) (2024). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

