TIMP1 Mediates Astrocyte-Dependent Local Immunosuppression in Brain Metastasis Acting on Infiltrating CD8+ T Cells
- PMID: 39354883
- PMCID: PMC11726018
- DOI: 10.1158/2159-8290.CD-24-0134
TIMP1 Mediates Astrocyte-Dependent Local Immunosuppression in Brain Metastasis Acting on Infiltrating CD8+ T Cells
Abstract
Immunotherapies against brain metastases have shown clinical benefits when applied to asymptomatic patients, but they are largely ineffective in symptomatic cases for unknown reasons. Here, we dissect the heterogeneity in metastasis-associated astrocytes using single-cell RNA sequencing and report a population that blocks the antitumoral activity of infiltrating T cells. This protumoral activity is mediated by the secretion of tissue inhibitor of metalloproteinase-1 (TIMP1) from a cluster of pSTAT3+ astrocytes that acts on CD63+ CD8+ T cells to modulate their function. Using genetic and pharmacologic approaches in mouse and human brain metastasis models, we demonstrate that combining immune checkpoint blockade antibodies with the inhibition of astrocyte-mediated local immunosuppression may benefit patients with symptomatic brain metastases. We further reveal that the presence of tissue inhibitor of metalloproteinase-1 in liquid biopsies provides a biomarker to select patients for this combined immunotherapy. Overall, our findings demonstrate an unexpected immunomodulatory role for astrocytes in brain metastases with clinical implications. Significance: This study presents a significant advancement in understanding immune modulation in brain tumors and offers new insights into the potential therapeutic interventions for brain metastases. See related commentary by Lorger and James, p. 11.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
N. Priego reports grants from Asociación Española Contra el Cáncer during the conduct of the study. A. de Pablos-Aragoneses reports grants from Fundación La Caixa during the conduct of the study. L. Álvaro-Espinosa reports grants from Spanish Ministry of Economy and Competitiveness during the conduct of the study. R. Rudà reports grants from Bayer, as well as personal fees from Novocure, Servier, Genenta, and CureVac outside the submitted work. M. Schmitz reports grants from Federal Ministry of Education and Research, cofunded by the European Commission, and the Federal Ministry of Education and Research during the conduct of the study. M. Valiente reports grants from AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Figures


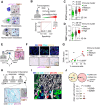
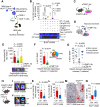

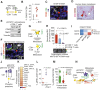
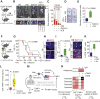
References
-
- Margolin K, Ernstoff MS, Hamid O, Lawrence D, McDermott D, Puzanov I, et al. . Ipilimumab in patients with melanoma and brain metastases: an open-label, phase 2 trial. Lancet Oncol 2012;13:459–65. - PubMed
-
- Gadgeel SM, Lukas RV, Goldschmidt J, Conkling P, Park K, Cortinovis D, et al. . Atezolizumab in patients with advanced non-small cell lung cancer and history of asymptomatic, treated brain metastases: exploratory analyses of the phase III OAK study. Lung Cancer 2019;128:105–12. - PubMed
MeSH terms
Substances
Grants and funding
- CIVP19S8163/Fundación Ramón Areces (FRA)
- RYC2022-038084-I/MICINN
- 4053/European Molecular Biology Organization (EMBO)
- R03 NS078392/NS/NINDS NIH HHS/United States
- 864759/HORIZON EUROPE European Research Council (ERC)
- 03ZU1111LB/Federal Ministry of Education and Research Germany
- NIH-NS078392/National Institutes of Health (NIH)
- LCF/BQ/DI19/11730044/"la Caixa" Foundation ("la Caixa")
- 01KT2304B/European Commission (EC)
- PID2019-107956RA-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- 141/Fundació la Marató de TV3 (Fundació la Marató)
- SAF2017-89643-R/Ministerio de Asuntos Económicos y Transformación Digital, Gobierno de España (MINECO)
- CIVP20A6613/Fundación Ramón Areces (FRA)
- AC20/00114/Instituto de Salud Carlos III (ISCIII)
- 54545/Cancer Research Institute (CRI)
- PRE_2019_1_0320/Eusko Jaurlaritza (Gobierno Vasco)
- BES-2017-081995/Ministerio de Asuntos Económicos y Transformación Digital, Gobierno de España (MINECO)
- LABAE211744PALA/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- TRANSCAN2021-2023/ERANET-TRANSCAN-3
- 828972/H2020-FETOPEN
- POSTD19016PRIE/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- RYC2018-024183-I/MICINN
- 804236/HORIZON EUROPE European Research Council (ERC)
- LABAE19002VALI/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- PRE2020-092342/Ministerio de Asuntos Económicos y Transformación Digital, Gobierno de España (MINECO)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

