Strategies for Making High-Performance Artificial Spider Silk Fibers
- PMID: 39355086
- PMCID: PMC11440630
- DOI: 10.1002/adfm.202305040
Strategies for Making High-Performance Artificial Spider Silk Fibers
Abstract
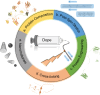
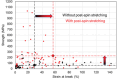
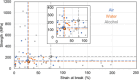
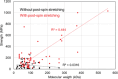
Artificial spider silk is an attractive material for many technical applications since it is a biobased fiber that can be produced under ambient conditions but still outcompetes synthetic fibers (e.g., Kevlar) in terms of toughness. Industrial use of this material requires bulk-scale production of recombinant spider silk proteins in heterologous host and replication of the pristine fiber's mechanical properties. High molecular weight spider silk proteins can be spun into fibers with impressive mechanical properties, but the production levels are too low to allow commercialization of the material. Small spider silk proteins, on the other hand, can be produced at yields that are compatible with industrial use, but the mechanical properties of such fibers need to be improved. Here, the literature on wet-spinning of artificial spider silk fibers is summarized and analyzed with a focus on mechanical performance. Furthermore, several strategies for how to improve the properties of such fibers, including optimized protein composition, smarter spinning setups, innovative protein engineering, chemical and physical crosslinking as well as the incorporation of nanomaterials in composite fibers, are outlined and discussed.
Keywords: biomimetic spinning; mechanical properties; protein fibers; rational designs; wet‐spinning.
© 2023 The Authors. Advanced Functional Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
