Individual variability in neural representations of mind-wandering
- PMID: 39355438
- PMCID: PMC11349032
- DOI: 10.1162/netn_a_00387
Individual variability in neural representations of mind-wandering
Abstract
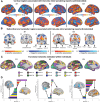
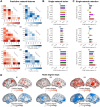
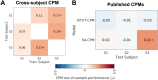
Mind-wandering is a frequent, daily mental activity, experienced in unique ways in each person. Yet neuroimaging evidence relating mind-wandering to brain activity, for example in the default mode network (DMN), has relied on population- rather than individual-based inferences owing to limited within-person sampling. Here, three densely sampled individuals each reported hundreds of mind-wandering episodes while undergoing multi-session functional magnetic resonance imaging. We found reliable associations between mind-wandering and DMN activation when estimating brain networks within individuals using precision functional mapping. However, the timing of spontaneous DMN activity relative to subjective reports, and the networks beyond DMN that were activated and deactivated during mind-wandering, were distinct across individuals. Connectome-based predictive modeling further revealed idiosyncratic, whole-brain functional connectivity patterns that consistently predicted mind-wandering within individuals but did not fully generalize across individuals. Predictive models of mind-wandering and attention that were derived from larger-scale neuroimaging datasets largely failed when applied to densely sampled individuals, further highlighting the need for personalized models. Our work offers novel evidence for both conserved and variable neural representations of self-reported mind-wandering in different individuals. The previously unrecognized interindividual variations reported here underscore the broader scientific value and potential clinical utility of idiographic approaches to brain-experience associations.
Keywords: Experience-sampling; Precision functional mapping; Resting-state fMRI; Spontaneous thought.
Plain language summary
While everyone experiences that their mind “wanders” throughout daily life, the content and form of inner experience is different in different people. In this study, we found that brain activity representing mind-wandering is different in each person, reflecting unique mental experiences. While people consistently engaged the brain’s default mode network (DMN) during mind-wandering, there were inconsistencies in the way that the DMN was engaged and in the other networks throughout the brain that were engaged. Our study highlights that personalized approaches, which require that individuals are sampled more densely than is common in current practice, enable accurate insights into relationships between brain activity and inner experience.
© 2024 Massachusetts Institute of Technology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures



Update of
-
Individual variability in neural representations of mind-wandering.bioRxiv [Preprint]. 2024 Jan 22:2024.01.20.576471. doi: 10.1101/2024.01.20.576471. bioRxiv. 2024. Update in: Netw Neurosci. 2024 Oct 01;8(3):808-836. doi: 10.1162/netn_a_00387. PMID: 38328109 Free PMC article. Updated. Preprint.
References
-
- Benisty, H., Barson, D., Moberly, A. H., Lohani, S., Tang, L., Coifman, R. R., Crair, M. C., Mishne, G., Cardin, J. A., & Higley, M. J. (2024). Rapid fluctuations in functional connectivity of cortical networks encode spontaneous behavior. Nature Neuroscience, 27(1), 148–158. 10.1038/s41593-023-01498-y, - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
