The genome sequence of a hydroid, Candelabrum cocksii (Cocks, 1854)
- PMID: 39355656
- PMCID: PMC11443189
- DOI: 10.12688/wellcomeopenres.22705.1
The genome sequence of a hydroid, Candelabrum cocksii (Cocks, 1854)
Abstract
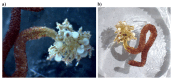
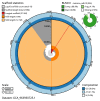
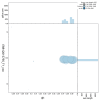
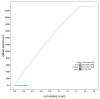
We present a genome assembly from an individual Candelabrum cocksii (hydroid; Cnidaria; Hydrozoa; Anthoathecata; Candelabridae). The genome sequence is 232.9 megabases in span. Most of the assembly is scaffolded into 15 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 14.55 kilobases in length.
Keywords: Anthoathecata; Candelabrum cocksii; a hydroid; chromosomal; genome sequence.
Copyright: © 2024 Adkins P et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
References
-
- Allman GJ: On the structure and development of Myriothela. Philosophical Transactions of the Royal Society of London (B). 1876;165:549–575. 10.1098/rstl.1875.0019 - DOI
LinkOut - more resources
Full Text Sources

