Targeting Immunoproteasome in Polarized Macrophages Ameliorates Experimental Emphysema Via Activating NRF1/2-P62 Axis and Suppressing IRF4 Transcription
- PMID: 39356034
- PMCID: PMC11600198
- DOI: 10.1002/advs.202405318
Targeting Immunoproteasome in Polarized Macrophages Ameliorates Experimental Emphysema Via Activating NRF1/2-P62 Axis and Suppressing IRF4 Transcription
Erratum in
-
Correction to "Targeting Immunoproteasome in Polarized Macrophages Ameliorates Experimental Emphysema via Activating NRF1/2-P62 Axis and Suppressing IRF4 Transcription".Adv Sci (Weinh). 2026 Jan 21:e24136. doi: 10.1002/advs.202524136. Online ahead of print. Adv Sci (Weinh). 2026. PMID: 41566951 No abstract available.
Abstract
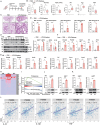
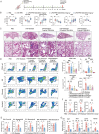
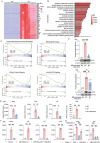
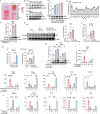
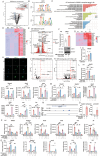
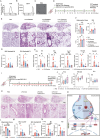
Chronic obstructive pulmonary disease (COPD) stands as the prevailing chronic airway ailment, characterized by chronic bronchitis and emphysema. Current medications fall short in treatment of these diseases, underscoring the urgent need for effective therapy. Prior research indicated immunoproteasome inhibition alleviated various inflammatory diseases by modulating immune cell functions. However, its therapeutic potential in COPD remains largely unexplored. Here, an elevated expression of immunoproteasome subunits LMP2 and LMP7 in the macrophages isolated from mouse with LPS/Elastase-induced emphysema and polarized macrophages in vitro is observed. Subsequently, intranasal administration of the immunoproteasome-specific inhibitor ONX-0914 significantly mitigated COPD-associated airway inflammation and improved lung function in mice by suppressing macrophage polarization. Additionally, ONX-0914 capsulated in PLGA nanoparticles exhibited more pronounced therapeutic effect on COPD than naked ONX-0914 by targeting immunoproteasome in polarized macrophages. Mechanistically, ONX-0914 activated autophagy and endoplasmic reticulum (ER) stress are not attribute to the ONX-0914 mediated suppression of macrophage polarization. Intriguingly, ONX-0914 inhibited M1 polarization through the nuclear factor erythroid 2-related factor-1 (NRF1) and NRF2-P62 axis, while the suppression of M2 polarization is regulated by inhibiting the transcription of interferon regulatory factor 4 (IRF4). In summary, the findings suggest that targeting immunoproteasome in macrophages holds promise as a therapeutic strategy for COPD.
Keywords: IRF4; NRF1/2‐P62; emphysema; immunoproteasome; macrophage polarization.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rabe K. F., Watz H., Lancet 2017, 389, 1931. - PubMed
-
- Brightling C., Greening N., Eur. Respir. J. 2019, 54. - PubMed
-
- Leitao Filho F. S., Takiguchi H., Akata K., Ra S. W., Moon J. Y., Kim H. K., Cho Y., Yamasaki K., Milne S., Yang J., Yang C. W. T., Li X., Nislow C., van Eeden S. F., Shaipanich T., Lam S., Leung J. M., Sin D. D., Am J. Respir. Crit. Care Med. 2021, 204, 1143. - PubMed
-
- Labaki W. W., Rosenberg S. R., Ann. Intern. Med. 2020, 173, ITC17. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFF0710802/National Key Technologies Research and Development Program of China
- 2022YFF0710800/National Key Technologies Research and Development Program of China
- 82470033/National Natural Science Foundation of China
- 32100734/National Natural Science Foundation of China
- 81803183/National Natural Science Foundation of China
- 32100914/National Natural Science Foundation of China
- 82200080/National Natural Science Foundation of China
- 2214050008970/Natural Science Foundation of Guangdong Province
- A2303030/Shenzhen Medical Research Fund
- B2302041/Shenzhen Medical Research Fund
- JCYJ20210324114400002/Shenzhen Science Technology and Innovative Commission (SZSTI)
- KCXFZ202002011008256/Shenzhen Science Technology and Innovative Commission (SZSTI)
- GantsC2302001/Shenzhen Medical Academy of Research and Translation
- JCYJ20210324114400001/Shenzhen Science and Technology Program
LinkOut - more resources
Full Text Sources
