USP36 SUMOylates Las1L and Promotes Its Function in Pre-Ribosomal RNA ITS2 Processing
- PMID: 39356143
- PMCID: PMC11523043
- DOI: 10.1158/2767-9764.CRC-24-0312
USP36 SUMOylates Las1L and Promotes Its Function in Pre-Ribosomal RNA ITS2 Processing
Abstract
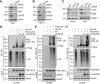
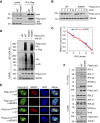
Ribosome biogenesis is a highly regulated cellular process requiring a large cohort of accessory factors to ensure the accurate production of ribosomes. Dysregulation of ribosome biogenesis is associated with the development of various human diseases, including cancer. The Las1L-Nol9 endonuclease-kinase complex is essential for the cleavage of the rRNA internal transcribed spacer 2 (ITS2), the phosphorylation of the 5'-hydroxyl end of the resulting precursor, and, thus, the maturation of the 60S ribosome. However, how the Las1L-Nol9 complex is regulated in cells is unclear. In this study, we report that the nucleolar ubiquitin-specific protease USP36 is a novel regulator of the Las1L-Nol9 complex. USP36 interacts with both Las1L and Nol9 and regulates their stability via deubiquitination. Intriguingly, USP36 also mediates the SUMOylation of Las1L, mainly at lysine (K) 565. Mutating K565 to arginine (R) does not affect the levels of Las1L and the formation of the Las1L-Nol9 complex, but abolishes its function in ITS2 processing, as unlike wild-type Las1L, the K565R mutant failed to rescue the defects in the ITS2 processing induced by the knockdown of endogenous Las1L. These results suggest that USP36-mediated Las1L SUMOylation is critical for ITS2 processing and that USP36 plays a critical role in ribosome biogenesis by regulating the Las1L-Nol9 complex.
Significance: This study identifies USP36 as a deubiquitinating and small ubiquitin-like modifier ligase dual-function enzyme to mediate Las1L deubiquitination and SUMOylation. Las1L SUMOylation at K565 plays a critical role in pre-rRNA ITS2 processing. Thus, our study reveals a novel downstream pathway for USP36-regulated ribosome biogenesis.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
R.C. Sears reports personal fees from Rappta Therapeutics and grants from AstraZeneca outside the submitted work. M.-S. Dai reports grants from NIH during the conduct of the study. No disclosures were reported by the other authors.
Figures





References
-
- Baßler J, Hurt E. Eukaryotic ribosome assembly. Annu Rev Biochem 2019;88:281–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

