Induction of the TEAD Coactivator VGLL1 by Estrogen Receptor-Targeted Therapy Drives Resistance in Breast Cancer
- PMID: 39356622
- PMCID: PMC7616691
- DOI: 10.1158/0008-5472.CAN-24-0013
Induction of the TEAD Coactivator VGLL1 by Estrogen Receptor-Targeted Therapy Drives Resistance in Breast Cancer
Abstract
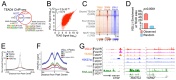
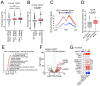
Resistance to endocrine therapies (ET) is common in estrogen receptor (ER)-positive breast cancer, and most relapsed patients die with ET-resistant disease. Although genetic mutations provide explanations for some relapses, mechanisms of resistance remain undefined in many cases. Drug-induced epigenetic reprogramming has been shown to provide possible routes to resistance. By analyzing histone H3 lysine 27 acetylation profiles and transcriptional reprogramming in models of ET resistance, we discovered that selective ER degraders, such as fulvestrant, promote expression of vestigial-like 1 (VGLL1), a coactivator for TEF-1 and AbaA domain (TEAD) transcription factors. VGLL1, acting via TEADs, promoted the expression of genes that drive the growth of fulvestrant-resistant breast cancer cells. Pharmacological disruption of VGLL1-TEAD4 interaction inhibited VGLL1/TEAD-induced transcriptional programs to prevent the growth of resistant cells. EGFR was among the VGLL1/TEAD-regulated genes, and VGLL1-directed EGFR upregulation sensitized fulvestrant-resistant breast cancer cells to EGFR inhibitors. Taken together, these findings identify VGLL1 as a transcriptional driver in ET resistance and advance therapeutic possibilities for relapsed ER+ breast cancer patients. Significance: Transcriptional reprogramming mediated by the upregulation of the TEAD coactivator VGLL1 confers resistance to estrogen receptor degraders in breast cancer but provides alternative therapeutic options for this clinically important patient group.
©2024 American Association for Cancer Research.
Conflict of interest statement
The authors declare no competing interests relevant to the work herein.
Figures




References
-
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. - PubMed
-
- Ali S, Buluwela L, Coombes RC. Antiestrogens and their therapeutic applications in breast cancer and other diseases. Annual review of medicine. 2011;62:217–32. - PubMed
-
- Johnston SR, Dowsett M. Aromatase inhibitors for breast cancer: lessons from the laboratory. Nature reviews Cancer. 2003;3:821–31. - PubMed
-
- Ma CX, Reinert T, Chmielewska I, Ellis MJ. Mechanisms of aromatase inhibitor resistance. Nature reviews Cancer. 2015;15:261–75. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

